The three-dimensional structure of the following target sequence has been predicted by YASARA's homology modeling experiment:
>T0454
MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQ
FACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPA
IASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDE
ELIRQYVAIFTRGIMADGGAAPA
The target sequence contains 203 residues in 1 molecule.
Not surprisingly, homology modeling involves a speed/accuracy trade-off. For this target, the requested speed was Slow, which implied the following parameters (some may have been set manually with the respective keywords):
Since the target sequence was the only available information, possible templates had to be identified by running PSI-BLAST to search the PDB for known structures with a similar sequence (i.e. hits with an E-value below the homology modeling cutoff 1.0).
The following 50 hits were found:
| Template | Total score | BLAST E-value | BLAST score | ID | Resolution | Header |
| 1 | 38.00 | 0.001 | 38.0 | 2OFL-B | 2.90 A | Chain B, Structural Genomics, The Crystal Structure Of A Tetr-Family Transcriptional Regulator From Streptomyces Coelicolor A3(2) |
| 2 | 35.56 | 5e-09 | 56.0 | 2JJ7-B | 2.10 A | Chain B, Crystal Structure Of The Hlyiir Mutant Protein With Residues 170-185 Substituted By Alanine (183 residues with quality score 0.635) |
| 3 | 32.90 | 3e-06 | 47.0 | 2FBQ-A | 1.80 A | Chain A, The Crystal Structure Of Transcriptional Regulator Pa3006 (213 residues with quality score 0.700) |
| 4 | 32.90 | 3e-06 | 47.0 | BFBQ-A | Unknown or NMR | This template is a re-refined version of the previous template, based on the structure factors deposited at the PDB. Trying both templates raises the chances for a good model. Visit the PDB-redo database [Joosten et al., www.cmbi.ru.nl/pdb_redo] for more details.To distinguish this template from the original, the first '2' has been replaced with 'B'. |
| 5 | 32.02 | 1e-04 | 41.0 | 2EH3-A | 1.55 A | Chain A, Crystal Structure Of Aq_1058, A Transcriptional Regulator (TerrACRR FAMILY) FROM AQUIFEX AEOLICUS VF5 (171 residues with quality score 0.781) |
| 6 | 30.96 | 0.0004 | 40.0 | 3BHQ-A | 1.54 A | Chain A, Crystal Structure Of Tetr-Family Transcriptional Regulator (Np_105615.1) From Mesorhizobium Loti At 1.54 A Resolution (202 residues with quality score 0.774) |
| 7 | 30.83 | 1e-08 | 54.0 | 2FX0-A | 2.40 A | Chain A, Crystal Structure Of Hlyiir, A Hemolysin Ii Transcriptional Regulator (179 residues with quality score 0.571) |
| 8 | 30.53 | 3e-08 | 53.0 | 1PB6-B | 2.50 A | Chain B, Crystal Structure Of Hypothetical Transcriptional Regulator Ycdc (198 residues with quality score 0.576) |
| 9 | 30.32 | 8e-05 | 42.0 | 2IBD-A | 1.50 A | Chain A, Crystal Structure Of Probable Transcriptional Regulatory Protein Rha5900 (192 residues with quality score 0.722) |
| 10 | 29.60 | 0.003 | 37.0 | 2IEK-A | 1.83 A | Chain A, New Crystal Form Of Transcriptional Regulator Tm1030 From Thermotoga Maritima (199 residues with quality score 0.800) |
| 10 | 29.25 | 0.0007 | 39.0 | 2QIB-A | - | Chain A, Crystal Structure Of Tetr-Family Transcriptional Regulator From Streptomyces Coelicolor (222 residues with quality score 0.750). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 28.53 | 0.002 | 37.0 | 2NP5-B | - | Chain B, Crystal Structure Of A Transcriptional Regulator (Rha1_ro04179) From Rhodococcus Sp. Rha1. (187 residues with quality score 0.771) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 28.40 | 0.005 | 36.0 | 2HYT-A | - | Chain A, Crystal Structure Of Tetr-Family Transcriptional Regulator (Yp_049917.1) From Erwinia Cartovora Atroseptica Scri1043 At 1.64 A Resolution (193 residues with quality score 0.789). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 28.28 | 0.0004 | 40.0 | 2GEN-A | - | Chain A, Structural Genomics, The Crystal Structure Of A Probable Transcriptional Regulator From Pseudomonas Aeruginosa Pao1 (188 residues with quality score 0.707). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 27.97 | 0.002 | 38.0 | 2ZCM-B | - | Chain B, Crystal Structure Of Icar, A Repressor Of The Tetr Family (180 residues with quality score 0.736). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 27.92 | 0.0003 | 40.0 | 2IAI-A | - | Chain A, Crystal Structure Of Sco3833, A Member Of The Tetr Transcriptional Regulator Family From Streptomyces Coelicolor A3 (201 residues with quality score 0.698). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 27.90 | 0.003 | 37.0 | 2ID6-A | - | Chain A, Crystal Structure Of Transcriptional Regulator (Tm1030) At 1.75a Resolution (202 residues with quality score 0.754). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 27.05 | 0.003 | 37.0 | 1Z77-A | - | Chain A, Crystal Structure Of Transcriptional Regulator Protein From Thermotoga Maritima. (200 residues with quality score 0.731). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 26.71 | 0.003 | 37.0 | 2ZCN-D | - | Chain D, Crystal Structure Of Icar, A Repressor Of The Tetr Family (184 residues with quality score 0.722) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 25.67 | 0.004 | 36.0 | 2G3B-B | - | Chain B, Crystal Structure Of Putative Tetr-Family Transcriptional Regulator From Rhodococcus Sp (188 residues with quality score 0.713). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 25.46 | 0.001 | 38.0 | 1T33-A | - | Chain A, Structural Genomics, The Crystal Structure Of A Putative Transcriptional Repressor (TetrACRR FAMILY) FROM Salmonella Typhimurim Lt2 (220 residues with quality score 0.670). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 25.01 | 0.003 | 37.0 | 1ZKG-B | - | Chain B, Crystal Structure Of Transcriptional Regulator, Tetr Family (Tm1030) From Thermotoga Maritima At 2.30 A Resolution (199 residues with quality score 0.676). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 24.98 | 0.037 | 33.0 | 2RAS-A | - | Chain A, Crystal Structure Of Predicted Transcriptional Regulator Of TetrACRR FAMILY (YP_495839.1) FROM NOVOSPHINGOBIUM Aromaticivorans Dsm 12444 At 1.80 A Resolution (200 residues with quality score 0.757). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 24.97 | 0.001 | 38.0 | 2DG8-A | - | Chain A, Crystal Structure Of The Putative Trasncriptional Regulator Sco7518 From Streptomyces Coelicolor A3(2) (175 residues with quality score 0.657) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 24.58 | 0.016 | 34.0 | 2QTQ-A | - | Chain A, Crystal Structure Of Predicted Dna-Binding Transcriptional Regulator (Yp_496351.1) From Novosphingobium Aromaticivorans Dsm 12444 At 1.85 A Resolution (204 residues with quality score 0.723) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 23.90 | 0.016 | 34.0 | 2RHA-A | - | Chain A, Crystal Structure Of Predicted Dna-Binding Transcriptional Regulator (Yp_496351.1) From Novosphingobium Aromaticivorans Dsm 12444 At 2.10 A Resolution (203 residues with quality score 0.703). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 23.87 | 0.006 | 36.0 | 1JT6-A | - | Chain A, Crystal Structure Of The Multidrug Binding Protein Qacr Bound To Dequalinium (186 residues with quality score 0.663) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 23.72 | 0.003 | 37.0 | 2D6Y-B | - | Chain B, Crystal Structure Of Transcriptional Factor Sco4008 From Streptomyces Coelicolor A3(2) (188 residues with quality score 0.641). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 23.43 | 0.048 | 33.0 | 2I10-B | - | Chain B, Putative Tetr Transcriptional Regulator From Rhodococcus Sp. Rha1 (182 residues with quality score 0.710). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 23.22 | 0.017 | 34.0 | 1UI5-A | - | Chain A, Crystal Structure Of Gamma-Butyrolactone Receptor (Arpa Like Protein) (199 residues with quality score 0.683). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 22.92 | 0.018 | 34.0 | 1UI6-A | - | Chain A, Crystal Structure Of Gamma-Butyrolactone Receptor (Arpa- Like Protein) (192 residues with quality score 0.674). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 22.42 | 0.004 | 37.0 | 2HKU-A | - | Chain A, Structural Genomics, The Crystal Structure Of A Putative Transcriptional Regulator From Rhodococcus Sp. Rha1 (191 residues with quality score 0.606). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 22.38 | 0.13 | 31.0 | 2REK-B | - | Chain B, Crystal Structure Of Tetr-Family Transcriptional Regulator (175 residues with quality score 0.722). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 22.10 | 0.023 | 34.0 | 1SGM-B | - | Chain B, Crystal Structure Of Hypothetical Protein Yxaf (184 residues with quality score 0.650). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 22.07 | 0.005 | 36.0 | 1JUS-A | - | Chain A, Crystal Structure Of The Multidrug Binding Transcriptional Repressor Qacr Bound To Rhodamine 6g (186 residues with quality score 0.613) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 21.42 | 0.004 | 36.0 | 2NP3-B | - | Chain B, Crystal Structure Of Tetr-Family Regulator (Sco0857) From Streptomyces Coelicolor A3 (180 residues with quality score 0.595). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 21.15 | 0.34 | 30.0 | 2FD5-A | - | Chain A, The Crystal Structure Of A Transcriptional Regulator From Pseudomonas Aeruginosa Pao1 (180 residues with quality score 0.705). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 20.99 | 0.006 | 36.0 | 1RKW-A | - | Chain A, Crystal Structure Of The Multidrug Binding Transcriptional Repressor Qacr Bound To Pentamadine (186 residues with quality score 0.583) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 20.70 | 0.063 | 32.0 | 2RAE-A | - | Chain A, Crystal Structure Of A TetrACRR FAMILY TRANSCRIPTIONAL Regulator From Rhodococcus Sp. Rha1 (193 residues with quality score 0.647). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 20.59 | 0.006 | 36.0 | 1JTX-A | - | Chain A, Crystal Structure Of The Multidrug Binding Transcriptional Regulator Qacr Bound To Crystal Violet (186 residues with quality score 0.572). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 20.05 | 0.006 | 36.0 | 1QVT-A | - | Chain A, Crystal Structure Of The Multidrug Binding Transcriptional Repressor Qacr Bound To The Drug Proflavine (186 residues with quality score 0.557) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 19.97 | 0.11 | 32.0 | 2QWT-A | - | Chain A, Crystal Structure Of The Tetr Transcription Regulatory Protein From Mycobacterium Vanbaalenii (167 residues with quality score 0.624). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 19.73 | 1e-06 | 48.0 | 2QOP-A | - | Chain A, Crystal Structure Of The Transcriptional Regulator Acrr From Escherichia Coli (207 residues with quality score 0.411) . NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 19.56 | 0.27 | 30.0 | 3CJD-B | - | Chain B, Crystal Structure Of Putative Tetr Transcriptional Regulator (Yp_510936.1) From Jannaschia Sp. Ccs1 At 1.79 A Resolution (182 residues with quality score 0.652). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 19.06 | 0.002 | 37.0 | 2PZ9-A | - | Chain A, Crystal Structure Of Putative Transcriptional Regulator Sco4942 From Streptomyces Coelicolor (179 residues with quality score 0.515). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 18.50 | 0.006 | 36.0 | 1QVU-A | - | Chain A, Crystal Structure Of The Multidrug Binding Transcriptional Repressor Qacr Bound To Two Drugs: Ethidium And Proflavine (186 residues with quality score 0.514). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 18.47 | 0.001 | 38.0 | 3C07-A | - | Chain A, Crystal Structure Of A Tetr Family Transcriptional Regulator From Streptomyces Coelicolor A3(2) (221 residues with quality score 0.486). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 17.37 | 0.02 | 34.0 | 2QCO-A | - | Chain A, Crystal Structure Of The Transcriptional Regulator Cmer From Campylobacter Jejuni (202 residues with quality score 0.511). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 16.47 | 0.53 | 29.0 | 2PBX-B | - | Chain B, Vibrio Cholerae Hapr (197 residues with quality score 0.568). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 16.08 | 1e-06 | 48.0 | 3BCG-B | - | Chain B, Conformational Changes Of The Acrr Regulator Reveal A Mechanism Of Induction (207 residues with quality score 0.335). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
| 10 | 14.66 | 0.099 | 32.0 | 3CDL-A | - | Chain A, Crystal Structure Of A Tetr Family Transcriptional Regulator From Pseudomonas Syringae Pv. Tomato Str. Dc3000 (187 residues with quality score 0.458). NOTE: This template was deliberately discarded: 10 templates with a higher score have already been found, and that is the requested maximum (use the 'Templates' keyword to change it). |
The 'Total score' in the second column is simply the product of the BLAST alignment score and the WHAT_CHECK [Hooft et al.,Nature 381:272] quality score in the PDBFinderII database. This makes sure that good template structures are used even if the alignment score is low. If no quality score has been determined yet in the PDBFinderII, a perfect score of 1.000 is assumed.
To aid alignment correction and loop modeling, a secondary structure prediction for the target sequence had to be obtained. This was achieved by running BLAST to retrieve homologous sequences, creating a multiple sequence alignment and feeding it to the 'Discrimination of Secondary structure Class (DSC)' prediction algorithm [King&Sternberg, Protein Sci. 5:2298-2310].
The resulting prediction is listed below, the lines 'PreHel', 'PreStr' and 'PreCoi' indicate the estimated prediction accuracy for the three secondary structure classes helix, strand and coil.
Sequence: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
SecStr : CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
PreHel : 11232555679998999999766432349999975323111111221124569999999999999999997653214797574445667854311557553232211111125766525699999999999997678979977555677999998555643468665565754212457999999999987621222211111
PreStr : 00111012110001000000000014560100000123778888410000010000000000000000001111000001323333221110100111335432100000011122210000001001000000001010011000000000001012136321000001121000000000000000011110121000000
PreCoi : 99767543321111111111344664201011135664221111479986531111111111111111112346896312213332222146699442222446799999974222375411110000111113431121122555433111111543331321445544235898653111111111112379767899999
For each of the templates listed above, models were built. Either a single model if the alignment was certain, or a number of alternative models if the alignment was ambiguous.
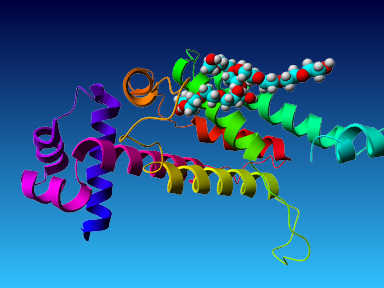
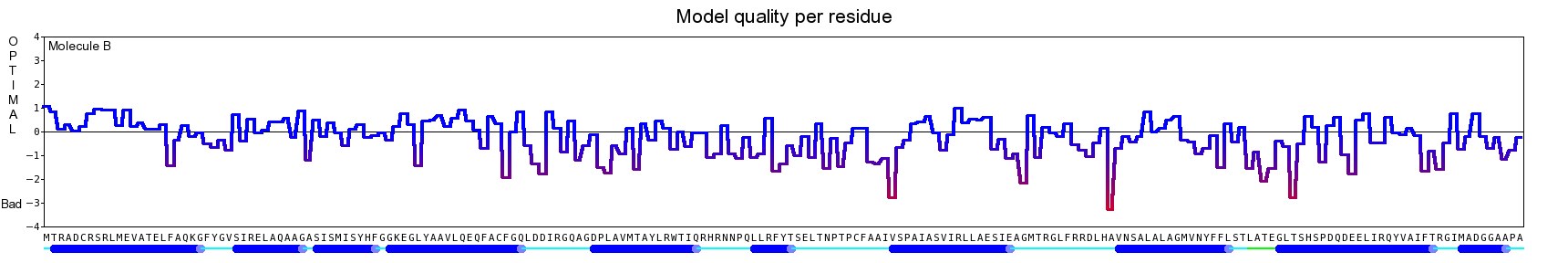
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||E:A :LF ||G| :::R |AQ AG:S|: Y|F:GKE L : :: A D A |:: L| |: H: Q::| : P F: : A :::I | | L: G|V Y:::: : : : |: :L : V:| ||
Template:.KSEQTRALILETAMRLFQERGYDRTTMRAIAQEAGVSVGNAYYYFAGKEHLIQGFYDRIAAEHRAAV.......DLEARLAGVLKVWLdYHEFAVQFFKNAA.....PLSPFSPESEHARVEAIGIHRAVLA..............ELMWLSQMGLVLYWIFDRTEGRERSYRLAERGARLTARGVVLARFRVLRPLVREVH
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHH.......HHHHHCHHHHHHHHHHHHHHHHHHHHHH.....CCTTTTCCHHHHHHHHHHHHHHHHH..............HHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHCTTTTTTTTHHHH
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRDLHA | VNSAL |
| 2 | FGQLD | DIRGQAG | DPLAV |
| 3 | RWTIQ | RHRN | NPQLL |
| 4 | LRFYT | SELTNPTPCFA | AIVSP |
| 5 | None | M | TRADC |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ofl-b01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -2.334 | Poor |
| Packing 1D | -3.011 | Bad |
| Packing 3D | -2.752 | Poor |
| Overall | -2.792 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ofl-b01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.599 | Good |
| Packing 1D | -1.571 | Satisfactory |
| Packing 3D | -0.660 | Good |
| Overall | -1.007 | Satisfactory |
Since the overall quality Z-score improved to -1.007 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 1 residue contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ofl-b01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 |  |
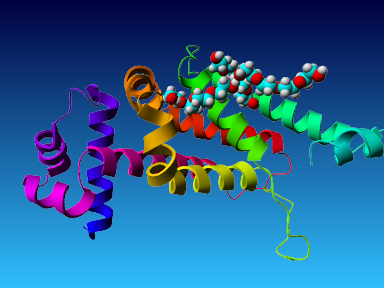
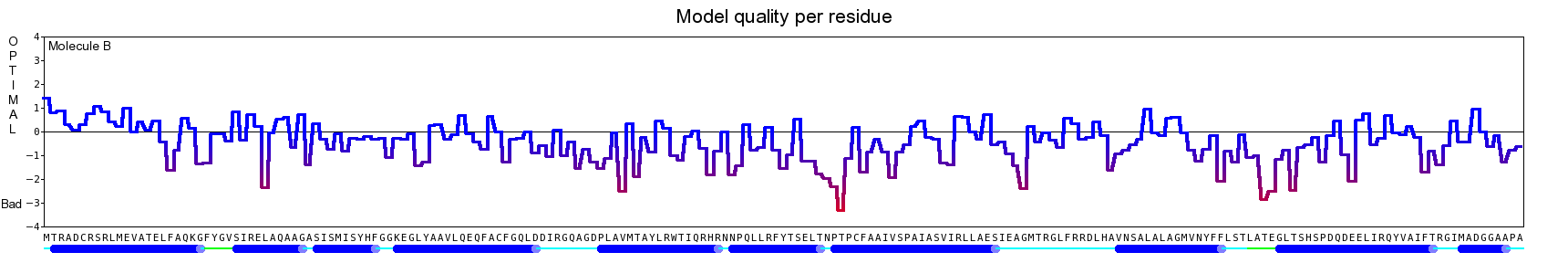
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||E:A :LF ||G| :::R |AQ AG:S|: Y|F:GKE L : :: A D A |:: L| |: |: | :: ::P F: : A :::I | | L: G|V Y:::: : : : |: :L : V:| ||
Template:.KSEQTRALILETAMRLFQERGYDRTTMRAIAQEAGVSVGNAYYYFAGKEHLIQGFYDRIAAEHRAAV.......DLEARLAGVLKVWLDIATPYHEFAVQFFKNAADP...FSPESEHARVEAIGIHRAVLA..............ELMWLSQMGLVLYWIFDRTEGRERSYRLAERGARLTARGVVLARFRVLRPLVREVH
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHH.......HHHHHCHHHHHHHHHHCCCHHHHHHHHHHHHHTT...TTCCHHHHHHHHHHHHHHHHH..............HHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHCTTTTTTTTHHHH
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRDLHA | VNSAL |
| 2 | FGQLD | DIRGQAG | DPLAV |
| 3 | SELTN | PTPCF | AAIVS |
| 4 | None | M | TRADC |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ofl-b02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -2.874 | Poor |
| Packing 1D | -3.193 | Bad |
| Packing 3D | -3.040 | Bad |
| Overall | -3.076 | Bad |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ofl-b02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.648 | Satisfactory |
| Packing 1D | -1.352 | Satisfactory |
| Packing 3D | -1.312 | Satisfactory |
| Overall | -1.376 | Satisfactory |
Since the overall quality Z-score improved to -1.376 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: The non-satisfactory model quality can have many reasons like limited structural similarity between template and target, alignment errors, bad template quality or large floppy loops. This model must be treated with extreme caution.
NOTE: 1 residue contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ofl-b02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 |  |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||E:A :LF ||G| :::R |AQ AG:S|: Y|F:GKE L : :: A D A |:: L| |: |: | :: ::P F: : A :::I | | RD| L: G|V Y:::: : : : |: :L : V:| :
Template:.KSEQTRALILETAMRLFQERGYDRTTMRAIAQEAGVSVGNAYYYFAGKEHLIQGFYDRIAAEHRAAV.......DLEARLAGVLKVWLDIATPYHEFAVQFFKNAADP...FSPESEHARVEAIGIHRAVLA.........RDILPELMWLSQMGLVLYWIFDRTEGRERSYRLAERGARLTARGVVLA..........REV
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHH.......HHHHHCHHHHHHHHHHCCCHHHHHHHHHHHHHTT...TTCCHHHHHHHHHHHHHHHHH.........CCHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHH..........HHH
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRR | DLHAV |
| 2 | FGQLD | DIRGQAG | DPLAV |
| 3 | YVAIF | TRGIMADGGA | APA |
| 4 | SELTN | PTPCF | AAIVS |
| 5 | None | M | TRADC |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ofl-b03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -3.062 | Bad |
| Packing 1D | -2.897 | Poor |
| Packing 3D | -2.871 | Poor |
| Overall | -2.909 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ofl-b03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.371 | Satisfactory |
| Packing 1D | -1.273 | Satisfactory |
| Packing 3D | -1.073 | Satisfactory |
| Overall | -1.194 | Satisfactory |
Since the overall quality Z-score improved to -1.194 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 1 residue contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ofl-b03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
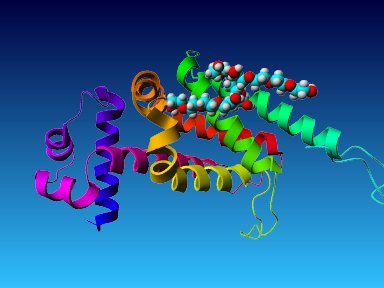 | 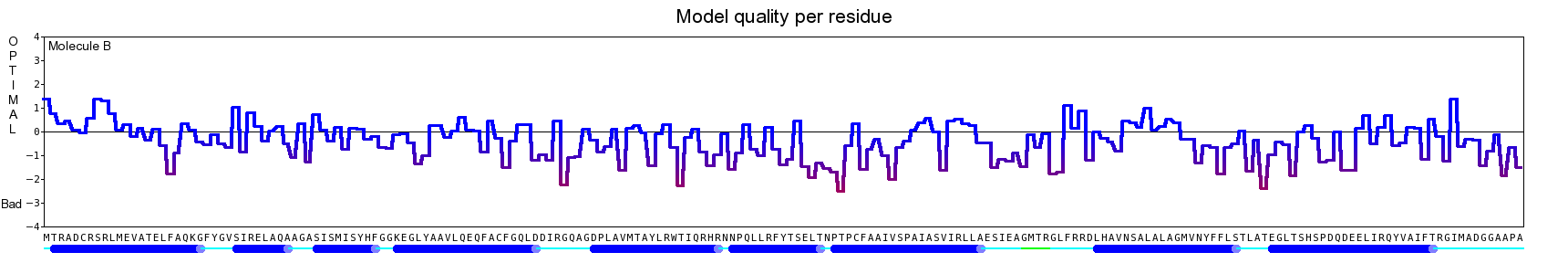 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||E:A :LF ||G| :::R |AQ AG:S|: Y|F:GKE L : :: A D A |:: L| |: H: Q::| : P F: : A :::I | | RD| L: G|V Y:::: : : : |: :L : V:| :
Template:.KSEQTRALILETAMRLFQERGYDRTTMRAIAQEAGVSVGNAYYYFAGKEHLIQGFYDRIAAEHRAAV.......DLEARLAGVLKVWLdYHEFAVQFFKNAA.....PLSPFSPESEHARVEAIGIHRAVLA.........RDILPELMWLSQMGLVLYWIFDRTEGRERSYRLAERGARLTARGVVLA........REVHE
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHH.......HHHHHCHHHHHHHHHHHHHHHHHHHHHH.....CCTTTTCCHHHHHHHHHHHHHHHHH.........CCHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHH........HHHHH
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRR | DLHAV |
| 2 | FGQLD | DIRGQAG | DPLAV |
| 3 | YVAIF | TRGIMADG | GAAPA |
| 4 | RWTIQ | RHRN | NPQLL |
| 5 | LRFYT | SELTNPTPCFA | AIVSP |
| 6 | None | M | TRADC |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ofl-b04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -2.484 | Poor |
| Packing 1D | -3.098 | Bad |
| Packing 3D | -2.732 | Poor |
| Overall | -2.839 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ofl-b04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.463 | Good |
| Packing 1D | -1.431 | Satisfactory |
| Packing 3D | -0.619 | Good |
| Overall | -0.913 | Good |
Since the overall quality Z-score improved to -0.913 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 1 residue contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ofl-b04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
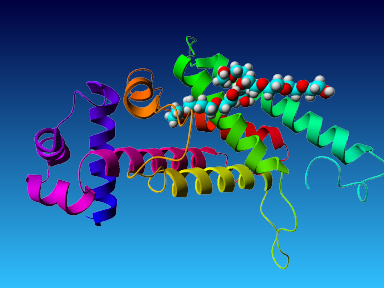 | 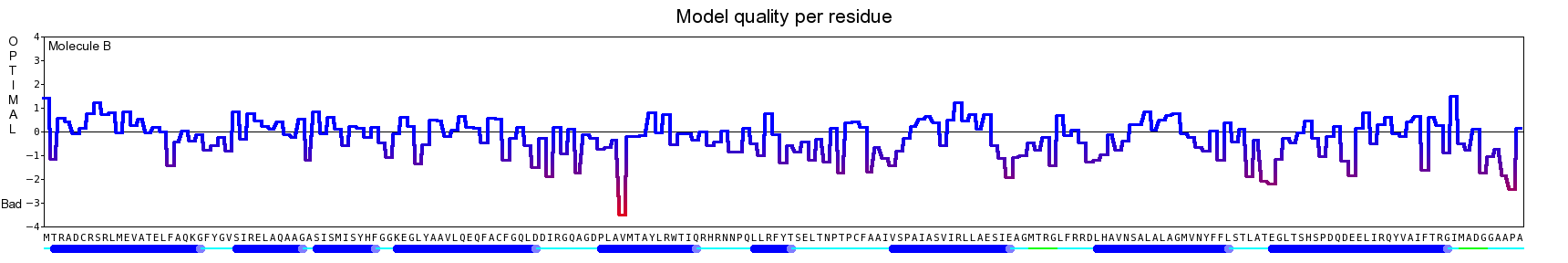 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||E:A :LF ||G| :::R |AQ AG:S|: Y|F:GKE L : :: A D A |:: L| |: |: | :: ::P F: : A :::I | | :: T RD| L: G|V Y:::: : : : |: :L : V:| :
Template:.KSEQTRALILETAMRLFQERGYDRTTMRAIAQEAGVSVGNAYYYFAGKEHLIQGFYDRIAAEHRAAV.......DLEARLAGVLKVWLDIATPYHEFAVQFFKNAADP...FSPESEHARVEAIGIHRAVLAGAKT.....RDILPELMWLSQMGLVLYWIFDRTEGRERSYRLAERGARLTARGVVLA........REVHE
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHH.......HHHHHCHHHHHHHHHHCCCHHHHHHHHHHHHHTT...TTCCHHHHHHHHHHHHHHHHHCCCC.....CCHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHH........HHHHH
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGLFRR | DLHAV |
| 2 | FGQLD | DIRGQAG | DPLAV |
| 3 | YVAIF | TRGIMADG | GAAPA |
| 4 | SELTN | PTPCF | AAIVS |
| 5 | None | M | TRADC |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ofl-b05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -3.271 | Bad |
| Packing 1D | -2.734 | Poor |
| Packing 3D | -2.999 | Poor |
| Overall | -2.935 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ofl-b05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.257 | Satisfactory |
| Packing 1D | -1.148 | Satisfactory |
| Packing 3D | -1.025 | Satisfactory |
| Overall | -1.107 | Satisfactory |
Since the overall quality Z-score improved to -1.107 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 1 residue contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ofl-b05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 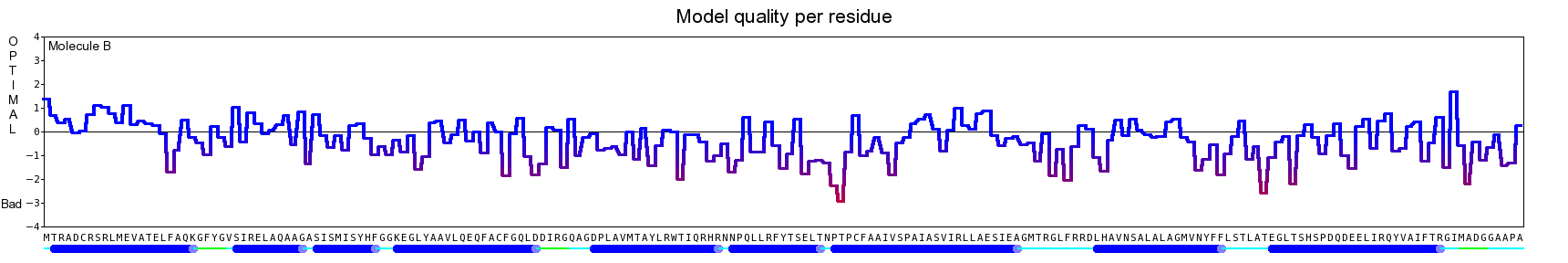 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| : |:P : : :: L E || G :G:F: : : :: F :: |L| : |: :T
Template:ASREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEIIK....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVLFPKF...............DSADLVSRIISALTD...........
SecStr: CHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHCHHHHHHHHHHHHHCCCCCCCHHHHHHCCHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHH....HHHHHHCCHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHTTTT...............HHHHHHHHHHHHHHH...........
The following 3 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | MVNYF | FLSTLATEGLTSHSPD | QDEEL |
| 2 | SELTN | PTPC | FAAIV |
| 3 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2jj7-b01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.148 | Satisfactory |
| Packing 1D | -1.719 | Satisfactory |
| Packing 3D | -1.254 | Satisfactory |
| Overall | -1.420 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2jj7-b01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.390 | Good |
| Packing 1D | -0.635 | Good |
| Packing 3D | -0.878 | Good |
| Overall | -0.712 | Good |
Since the overall quality Z-score improved to -0.712 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 11 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2jj7-b01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 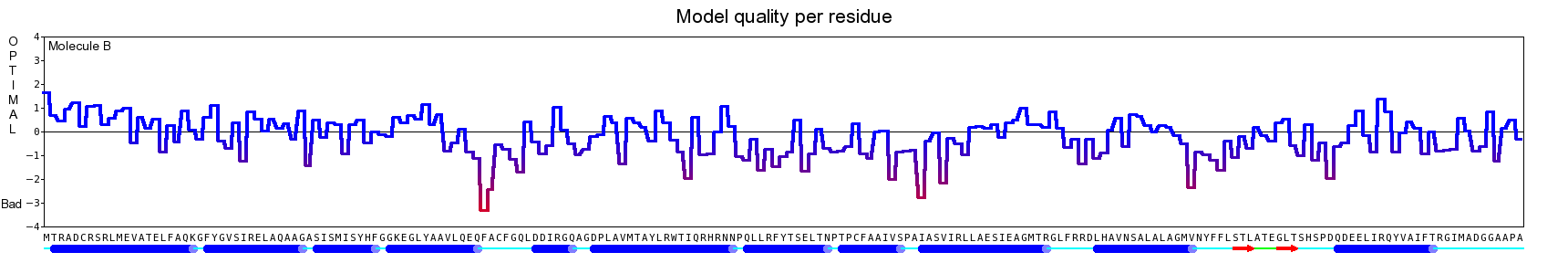 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| : : : I : : :: L E || G :G:F: : : :: F :: |L| : |: :T
Template:ASREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEIIKESARLEKIK...FIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVLFPKF...............DSADLVSRIISALTD...........
SecStr: CHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHCCHHHHHHHHHCHHHHHHHHHHHHHCCCCCCCHHHHHHCCHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHCCCHHHHHH...HHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHTTTT...............HHHHHHHHHHHHHHH...........
The following 3 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | MVNYF | FLSTLATEGLTSHSPD | QDEEL |
| 2 | FAAIV | SPA | IASVI |
| 3 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2jj7-b02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.448 | Satisfactory |
| Packing 1D | -1.436 | Satisfactory |
| Packing 3D | -1.291 | Satisfactory |
| Overall | -1.370 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2jj7-b02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.902 | Good |
| Packing 1D | -0.415 | Good |
| Packing 3D | -0.843 | Good |
| Overall | -0.685 | Good |
Since the overall quality Z-score improved to -0.685 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 11 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
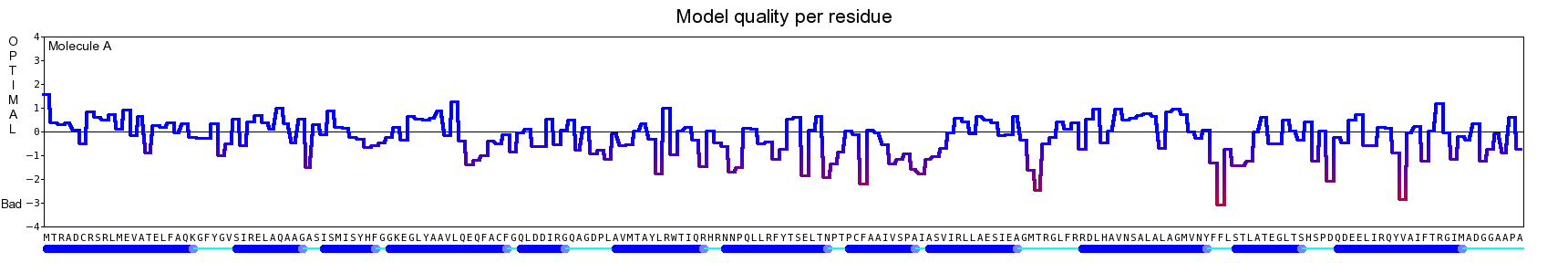
The final model for this template and alignment has been saved as t0454_2jj7-b02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 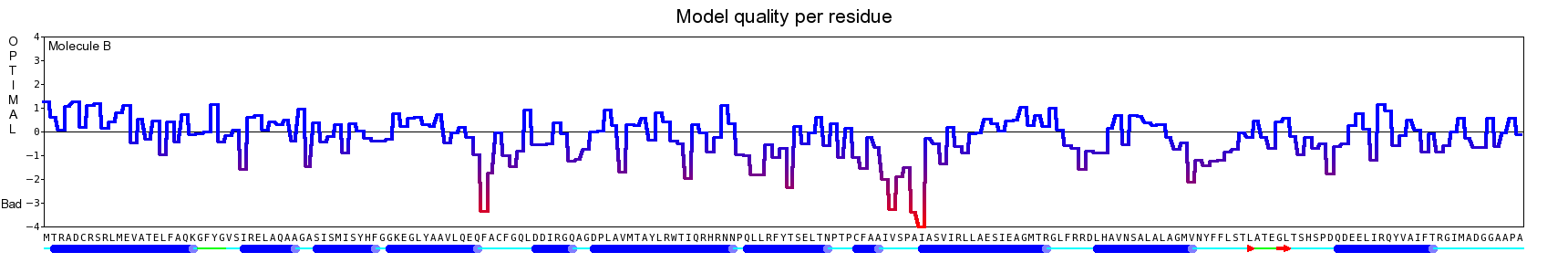 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : |: : | G : | : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA......PPIELFWRVHFMLGAAAFsIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH......HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGLFR | RDLHA |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | MVNYF | FL | STLAT |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fbq-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.144 | Satisfactory |
| Packing 1D | -1.781 | Satisfactory |
| Packing 3D | -0.827 | Good |
| Overall | -1.246 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fbq-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.571 | Good |
| Packing 1D | -0.988 | Good |
| Packing 3D | -0.632 | Good |
| Overall | -0.762 | Good |
Since the overall quality Z-score improved to -0.762 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
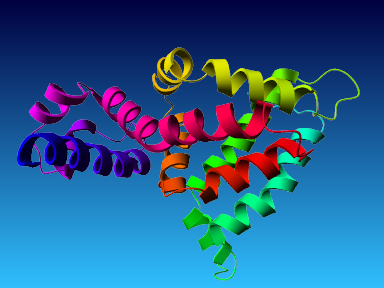
The final model for this template and alignment has been saved as t0454_2fbq-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
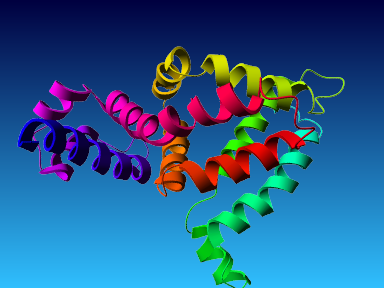 |  |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : |: : | G : | : : A ::|:: : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA......PPIELFWRVHFMLGAAAFSMSGIKALRAMAE....STEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH......HHHHHHHHHHHHHHHHHHHHHCHHHHHHHHH....HHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGLFR | RDLHA |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | EGLTS | HSPD | QDEEL |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fbq-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.364 | Satisfactory |
| Packing 1D | -1.586 | Satisfactory |
| Packing 3D | -0.795 | Good |
| Overall | -1.186 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fbq-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.579 | Good |
| Packing 1D | -0.884 | Good |
| Packing 3D | -0.538 | Good |
| Overall | -0.679 | Good |
Since the overall quality Z-score improved to -0.679 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fbq-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 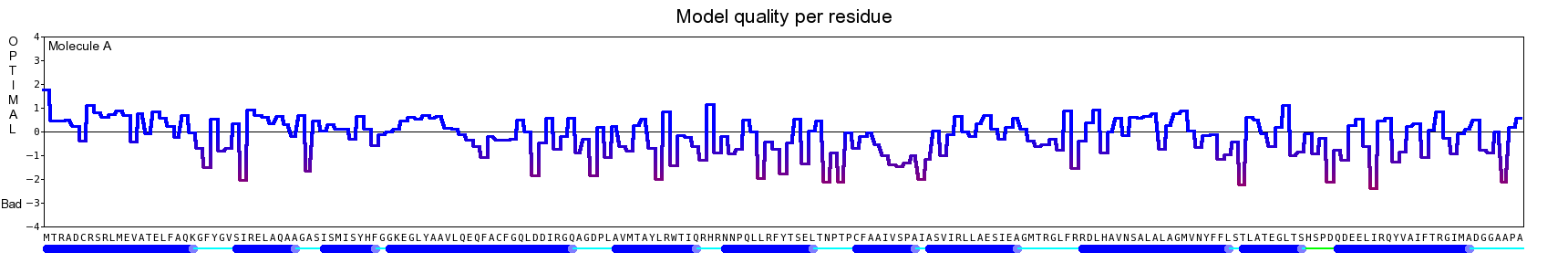 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |L | A: : |::L : : T: : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIELFWRVHFMLGAAAFSMSGIKALRAMAETDF...STEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHH...HHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | GLTSH | SPD | QDEEL |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fbq-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.300 | Satisfactory |
| Packing 1D | -1.435 | Satisfactory |
| Packing 3D | -0.667 | Good |
| Overall | -1.059 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fbq-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.700 | Good |
| Packing 1D | -0.671 | Good |
| Packing 3D | -0.461 | Good |
| Overall | -0.578 | Good |
Since the overall quality Z-score improved to -0.578 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fbq-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
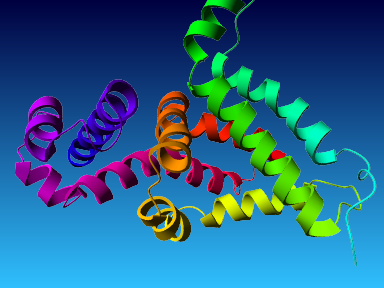 | 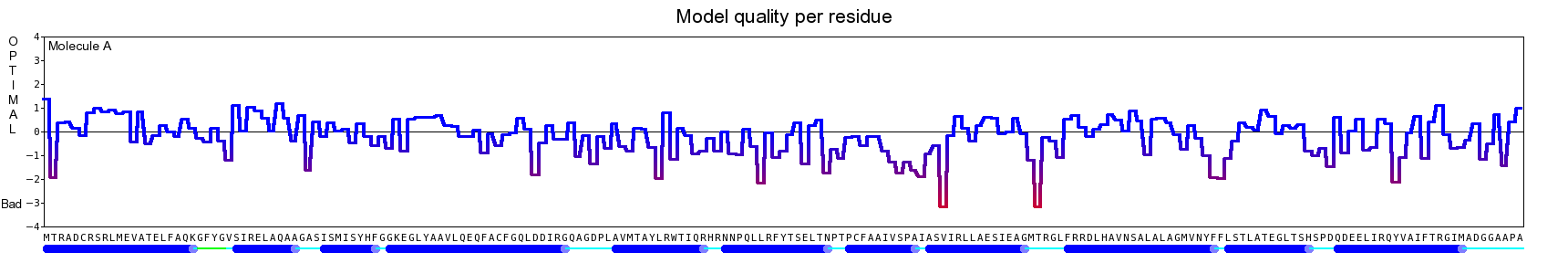 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |L | A: : : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIELFWRVHFMLGAAAFSMsIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | MVNYF | FL | STLAT |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fbq-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.084 | Satisfactory |
| Packing 1D | -1.826 | Satisfactory |
| Packing 3D | -0.711 | Good |
| Overall | -1.201 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fbq-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.428 | Good |
| Packing 1D | -1.031 | Satisfactory |
| Packing 3D | -0.539 | Good |
| Overall | -0.715 | Good |
Since the overall quality Z-score improved to -0.715 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fbq-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
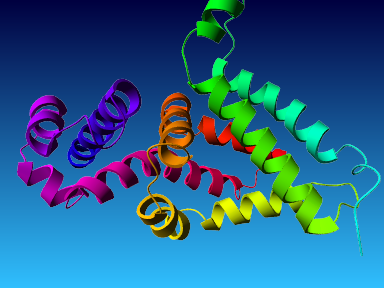 | 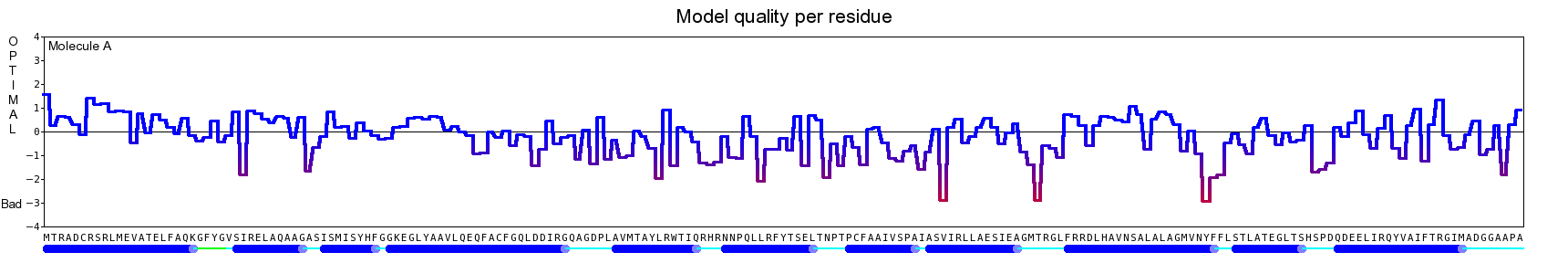 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |: V: L: A:: : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIeFWRVHFMLGAAAFSMSGIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | GLFRR | D | LHAVN |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fbq-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.351 | Satisfactory |
| Packing 1D | -1.770 | Satisfactory |
| Packing 3D | -0.754 | Good |
| Overall | -1.238 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fbq-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.506 | Good |
| Packing 1D | -1.118 | Satisfactory |
| Packing 3D | -0.631 | Good |
| Overall | -0.803 | Good |
Since the overall quality Z-score improved to -0.803 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fbq-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
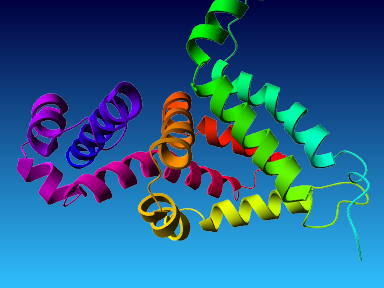 | 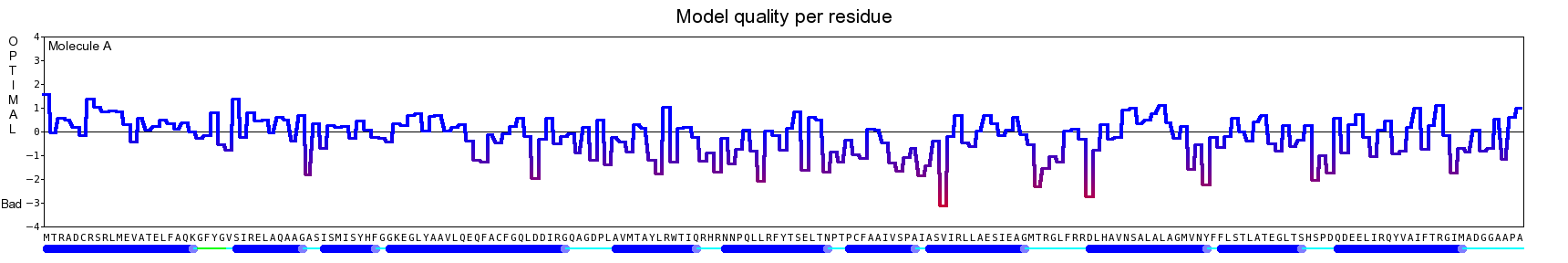 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : |: : | G : | : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA......PPIELFWRVHFMLGAAAFsIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH......HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGLFR | RDLHA |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | MVNYF | FL | STLAT |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_bfbq-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.337 | Satisfactory |
| Packing 1D | -1.673 | Satisfactory |
| Packing 3D | -0.853 | Good |
| Overall | -1.243 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_bfbq-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.654 | Good |
| Packing 1D | -0.792 | Good |
| Packing 3D | -0.551 | Good |
| Overall | -0.660 | Good |
Since the overall quality Z-score improved to -0.660 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_bfbq-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
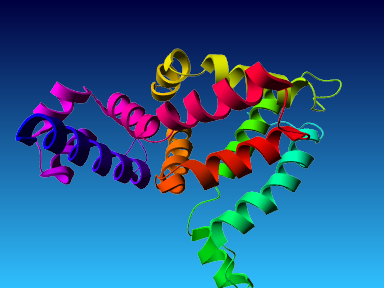 | 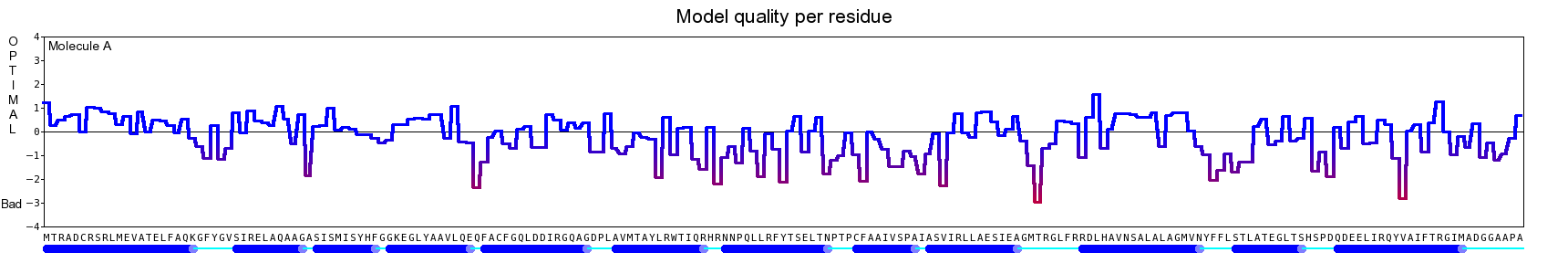 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : |: : | G : | : : A ::|:: : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA......PPIELFWRVHFMLGAAAFSMSGIKALRAMAE....STEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH......HHHHHHHHHHHHHHHHHHHHHCHHHHHHHHH....HHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGLFR | RDLHA |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | EGLTS | HSPD | QDEEL |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_bfbq-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.226 | Satisfactory |
| Packing 1D | -1.387 | Satisfactory |
| Packing 3D | -0.664 | Good |
| Overall | -1.028 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_bfbq-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.657 | Good |
| Packing 1D | -0.585 | Good |
| Packing 3D | -0.449 | Good |
| Overall | -0.532 | Good |
Since the overall quality Z-score improved to -0.532 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_bfbq-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 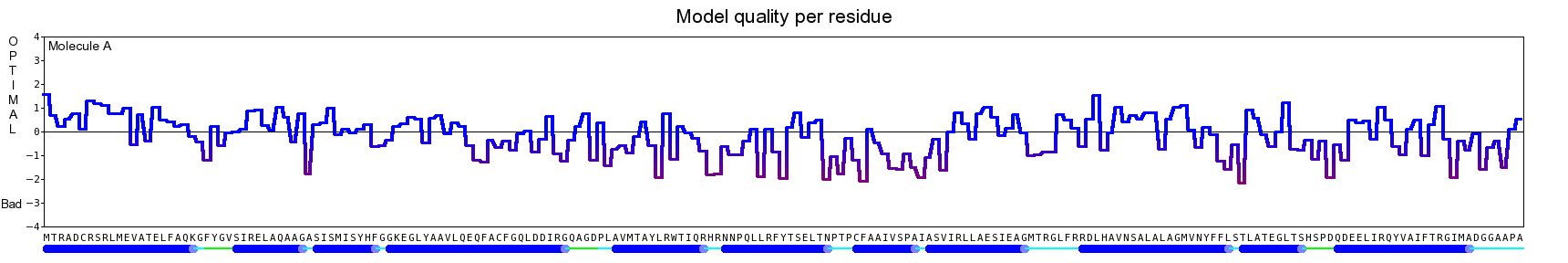 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |L | A: : |::L : : T: : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIELFWRVHFMLGAAAFSMSGIKALRAMAETDF...STEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHH...HHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | GLTSH | SPD | QDEEL |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_bfbq-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.373 | Satisfactory |
| Packing 1D | -1.417 | Satisfactory |
| Packing 3D | -0.583 | Good |
| Overall | -1.024 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_bfbq-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.469 | Good |
| Packing 1D | -0.500 | Good |
| Packing 3D | -0.148 | Good |
| Overall | -0.332 | Good |
Since the overall quality Z-score improved to -0.332 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_bfbq-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 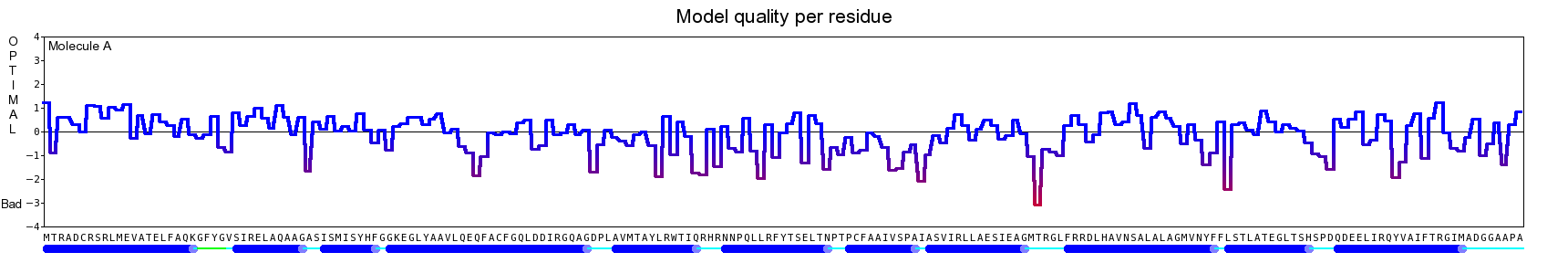 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |L | A: : : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIELFWRVHFMLGAAAFSMsIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | MVNYF | FL | STLAT |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_bfbq-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.247 | Satisfactory |
| Packing 1D | -1.504 | Satisfactory |
| Packing 3D | -0.655 | Good |
| Overall | -1.072 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_bfbq-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.524 | Good |
| Packing 1D | -0.814 | Good |
| Packing 3D | -0.503 | Good |
| Overall | -0.627 | Good |
Since the overall quality Z-score improved to -0.627 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_bfbq-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
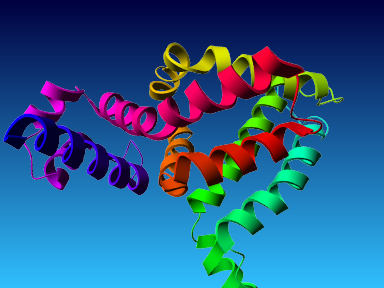 | 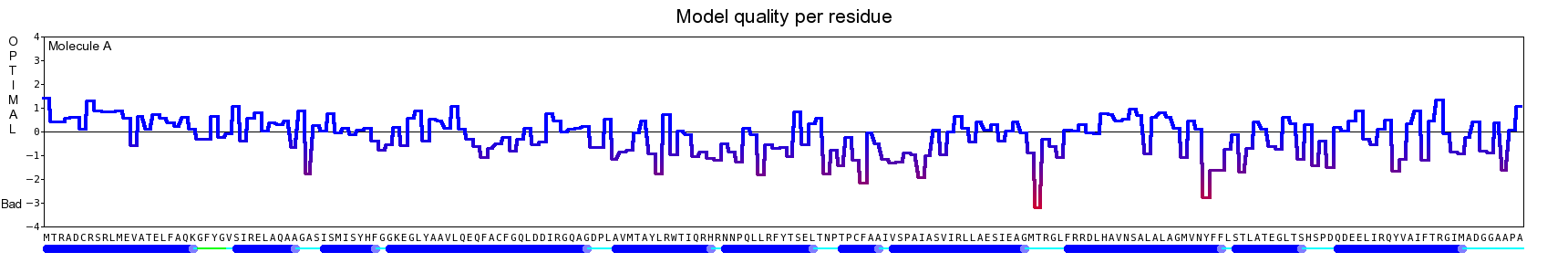 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::| :R|||:A |LFA|KGF :S|R |:: AG::|: |:YHFG:K::L AV::: :: F | : :| : :: :: :|R: ::: : :: : ::V:R |: : L |: V: L: A:: : : A : E|::: V :F: G: A|:G
Template:.AQSETVERILDAAEQLFAEKGFAETSLRLITSKAGVNLAAVNYHFGSKKALIQAVFSRFLGPFCASLEKEL......DLLHLLVSQAMA..NDLSIFMRLLGLAFSQSQGHLRKYLEEVYGKVFRRYMLLVNEAA...LPPIeFWRVHFMLGAAAFSMSGIKALRAMAETDFGVNTSTEQVMHLMVPFFAAGMRAESG....
SecStr: .HHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH......HHHHHHHHHHHH..HHHHHHHHHHHHHHHHCHHHHHHHHHHHCHHHHHHHHHHHHHHH...CHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHCCCC....
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | IEAGM | TRGL | FRRDL |
| 2 | DDIRG | QAGDPL | AVMTA |
| 3 | RWTIQ | RHR | NNPQL |
| 4 | GLFRR | D | LHAVN |
| 5 | None | M | TRADC |
| 6 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_bfbq-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.077 | Satisfactory |
| Packing 1D | -1.729 | Satisfactory |
| Packing 3D | -0.612 | Good |
| Overall | -1.116 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_bfbq-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.421 | Good |
| Packing 1D | -1.023 | Satisfactory |
| Packing 3D | -0.558 | Good |
| Overall | -0.720 | Good |
Since the overall quality Z-score improved to -0.720 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 5 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_bfbq-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
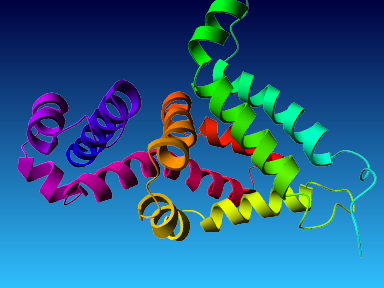 | 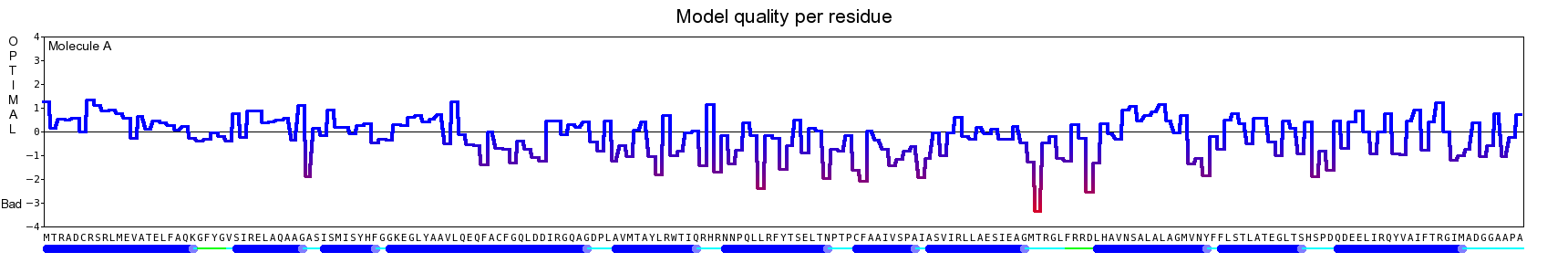 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |:R||EV: ELF |KG| G:S|:E|:: A: S : : |HF :KE L : |||: :: :|| P :| |L: : ::: :| |L F I I :: R : : |E | S | L: | : :: :| E : P E:L : IF
Template:....GTKERILEVSKELFFEKGYQGTSVEEIVKRANLSKGAFYFHFKSKEELITEIIERTHkIISLFEEN.....TPEELLEMFLEVLYR.....KKVVYIFLFDLL.....FRNIYFEKIEDAKRRFEKFLE...........KAEILSEIILGFLRQLILHYVIKEE....ELPFLKEKLREGLKLIF.............
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHH.....HHHHHHHHHHHHHHH.....HHHHHHHHHHHH.....HHHHHHHHHHHHHHHHHHHHH...........HHHHHHHHHHHHHHHHHHHHHHCCC....HHHHHHHHHHHHHHHHH.............
The following 8 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRD | LHAVN |
| 2 | STLAT | EGLTS | HSPDQ |
| 3 | QLDDI | RGQAG | DPLAV |
| 4 | YVAIF | TRGIMADGGAAPA | None |
| 5 | RWTIQ | RHRNN | PQLLR |
| 6 | LQEQF | A | CFGQL |
| 7 | TSELT | NPTPC | FAAIV |
| 8 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2eh3-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.240 | Satisfactory |
| Packing 1D | -1.221 | Satisfactory |
| Packing 3D | -0.513 | Good |
| Overall | -0.895 | Good |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2eh3-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.729 | Good |
| Packing 1D | -0.581 | Good |
| Packing 3D | -0.464 | Good |
| Overall | -0.548 | Good |
Since the overall quality Z-score improved to -0.548 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 17 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2eh3-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 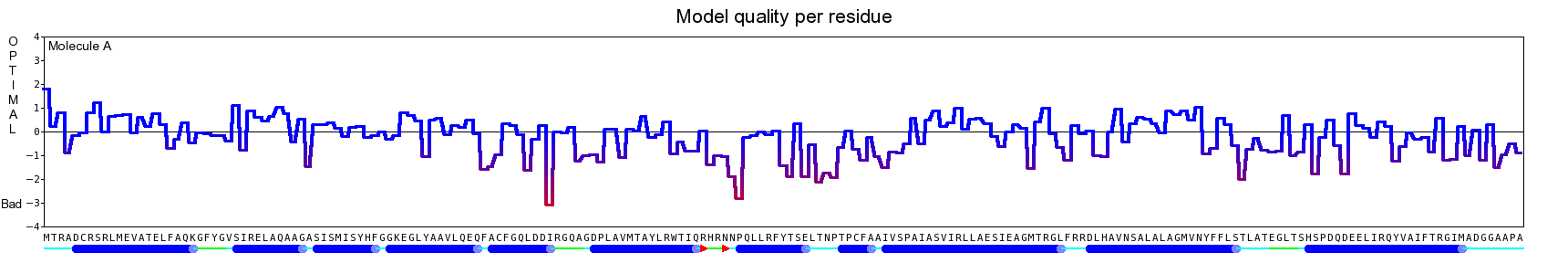 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |:R||EV: ELF |KG| G:S|:E|:: A: S : : |HF :KE L : |||: :: :|| P :| |L: :| L: : SE F I I :: R : : |E | S | L: | : :: :| E : P E:L : IF
Template:....GTKERILEVSKELFFEKGYQGTSVEEIVKRANLSKGAFYFHFKSKEELITEIIERTHkIISLFEEN.....TPEELLEMFLEVLYrKKVVYIFLFDLLCSE.......FRNIYFEKIEDAKRRFEKFLE...........KAEILSEIILGFLRQLILHYVIKEE....ELPFLKEKLREGLKLIF.............
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHH.....HHHHHHHHHHHHHHHHHHHHHHHHHHHCCC.......HHHHHHHHHHHHHHHHHHHHH...........HHHHHHHHHHHHHHHHHHHHHHCCC....HHHHHHHHHHHHHHHHH.............
The following 8 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRD | LHAVN |
| 2 | STLAT | EGLTS | HSPDQ |
| 3 | QLDDI | RGQAG | DPLAV |
| 4 | YVAIF | TRGIMADGGAAPA | None |
| 5 | LQEQF | A | CFGQL |
| 6 | RWTIQ | RH | RNNPQ |
| 7 | LRFYT | SELTNPTPC | FAAIV |
| 8 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2eh3-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.062 | Satisfactory |
| Packing 1D | -1.836 | Satisfactory |
| Packing 3D | -0.806 | Good |
| Overall | -1.246 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2eh3-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.727 | Good |
| Packing 1D | -0.968 | Good |
| Packing 3D | -0.800 | Good |
| Overall | -0.855 | Good |
Since the overall quality Z-score improved to -0.855 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 17 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2eh3-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 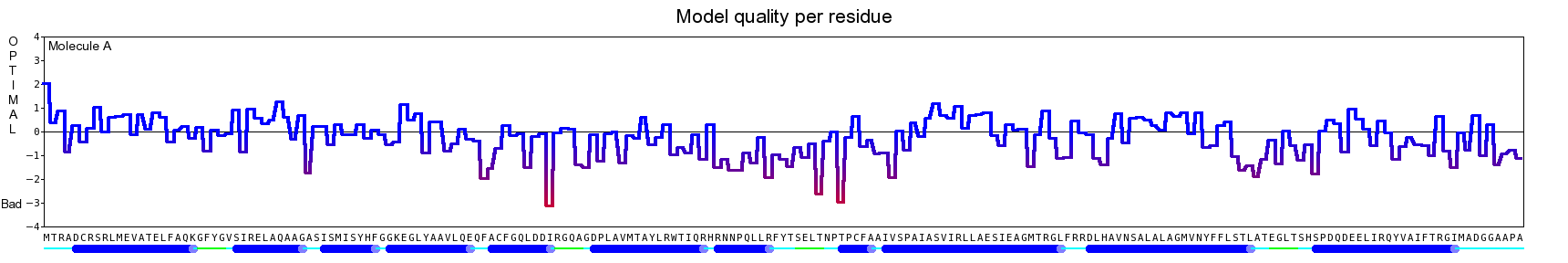 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |:R||EV: ELF |KG| G:S|:E|:: A: S : : |HF :KE L : |||: :: :|| P :| |L: : : : F : L F I I :: R : : |E | S | L: | : :: :| E : P E:L : IF
Template:....GTKERILEVSKELFFEKGYQGTSVEEIVKRANLSKGAFYFHFKSKEELITEIIERTHkIISLFEEN.....TPEELLEMFLEVLYR....KKVVYIFLFDLL......FRNIYFEKIEDAKRRFEKFLE...........KAEILSEIILGFLRQLILHYVIKEE....ELPFLKEKLREGLKLIF.............
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHH.....HHHHHHHHHHHHHHH....HHHHHHHHHHHH......HHHHHHHHHHHHHHHHHHHHH...........HHHHHHHHHHHHHHHHHHHHHHCCC....HHHHHHHHHHHHHHHHH.............
The following 8 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRD | LHAVN |
| 2 | STLAT | EGLTS | HSPDQ |
| 3 | QLDDI | RGQAG | DPLAV |
| 4 | YVAIF | TRGIMADGGAAPA | None |
| 5 | RWTIQ | RHRN | NPQLL |
| 6 | LQEQF | A | CFGQL |
| 7 | YTSEL | TNPTPC | FAAIV |
| 8 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2eh3-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.021 | Satisfactory |
| Packing 1D | -1.684 | Satisfactory |
| Packing 3D | -0.585 | Good |
| Overall | -1.077 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2eh3-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.865 | Good |
| Packing 1D | -0.869 | Good |
| Packing 3D | -0.675 | Good |
| Overall | -0.778 | Good |
Since the overall quality Z-score improved to -0.778 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 17 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2eh3-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
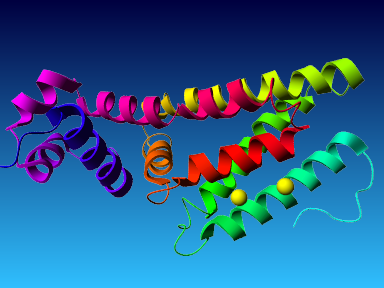 | 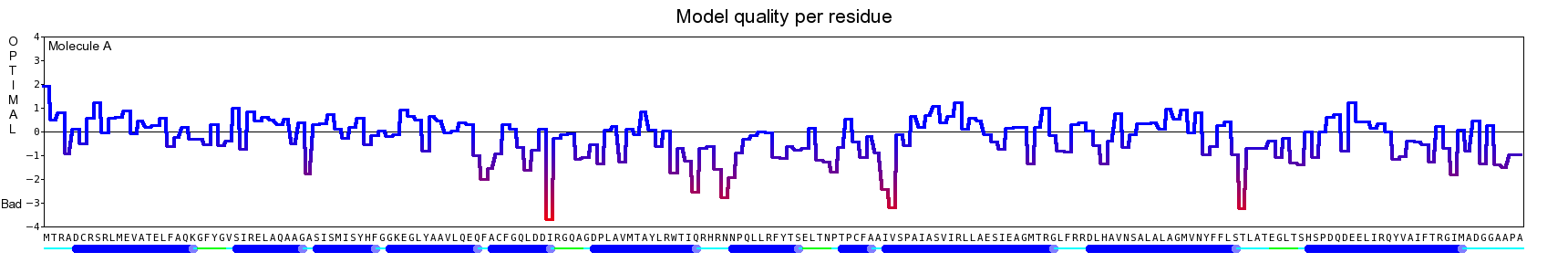 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |:R||EV: ELF |KG| G:S|:E|:: A: S : : |HF :KE L : |||: :: :|| P :| |L: : ::: :| |L F I I :: R : : |E | S | | G::: ::L : E : P E:L : IF
Template:....GTKERILEVSKELFFEKGYQGTSVEEIVKRANLSKGAFYFHFKSKEELITEIIERTHkIISLFEEN.....TPEELLEMFLEVLYR.....KKVVYIFLFDLL.....FRNIYFEKIEDAKRRFEKFLE...........KAEILSEI.ILGFLRQLILHYVIKE....ELPFLKEKLREGLKLIF.............
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHH.....HHHHHHHHHHHHHHH.....HHHHHHHHHHHH.....HHHHHHHHHHHHHHHHHHHHH...........HHHHHHHH.HHHHHHHHHHHHHHCC....HHHHHHHHHHHHHHHHH.............
The following 9 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRD | LHAVN |
| 2 | TLATE | GLTS | HSPDQ |
| 3 | QLDDI | RGQAG | DPLAV |
| 4 | YVAIF | TRGIMADGGAAPA | None |
| 5 | AVNSA | LA | LAGMV |
| 6 | RWTIQ | RHRNN | PQLLR |
| 7 | LQEQF | A | CFGQL |
| 8 | TSELT | NPTPC | FAAIV |
| 9 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2eh3-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.367 | Satisfactory |
| Packing 1D | -1.540 | Satisfactory |
| Packing 3D | -0.781 | Good |
| Overall | -1.162 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2eh3-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.966 | Good |
| Packing 1D | -0.743 | Good |
| Packing 3D | -0.721 | Good |
| Overall | -0.765 | Good |
Since the overall quality Z-score improved to -0.765 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 17 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2eh3-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
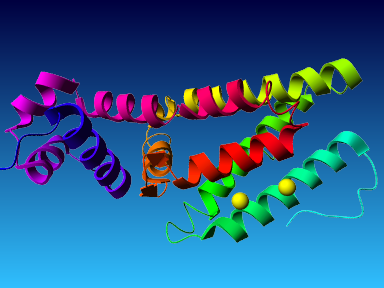 | 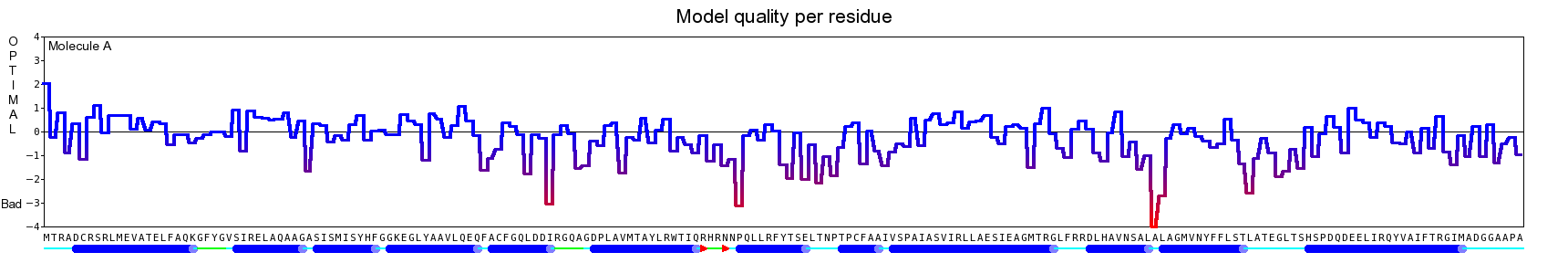 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |:R||EV: ELF |KG| G:S|:E|:: A: S : : |HF :KE L : |||: : :L | : P :| |L: : ::: :| |L F I I :: R : : |E | S | L: | : :: :| E : P E:L : IF
Template:....GTKERILEVSKELFFEKGYQGTSVEEIVKRANLSKGAFYFHFKSKEELITEIIERTHKKIISLFEEN....TPEELLEMFLEVLYR.....KKVVYIFLFDLL.....FRNIYFEKIEDAKRRFEKFLE...........KAEILSEIILGFLRQLILHYVIKEE....ELPFLKEKLREGLKLIF.............
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHH....HHHHHHHHHHHHHHH.....HHHHHHHHHHHH.....HHHHHHHHHHHHHHHHHHHHH...........HHHHHHHHHHHHHHHHHHHHHHCCC....HHHHHHHHHHHHHHHHH.............
The following 7 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AESIE | AGMTRGLFRRD | LHAVN |
| 2 | STLAT | EGLTS | HSPDQ |
| 3 | LDDIR | GQAG | DPLAV |
| 4 | YVAIF | TRGIMADGGAAPA | None |
| 5 | RWTIQ | RHRNN | PQLLR |
| 6 | TSELT | NPTPC | FAAIV |
| 7 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2eh3-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.033 | Satisfactory |
| Packing 1D | -1.124 | Satisfactory |
| Packing 3D | -0.383 | Good |
| Overall | -0.767 | Good |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2eh3-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.591 | Good |
| Packing 1D | -0.707 | Good |
| Packing 3D | -0.512 | Good |
| Overall | -0.600 | Good |
Since the overall quality Z-score improved to -0.600 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 17 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2eh3-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
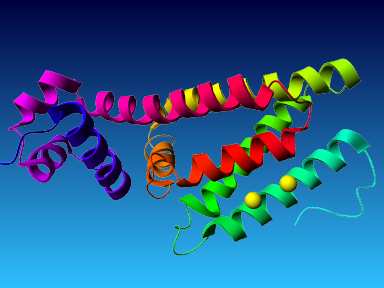 | 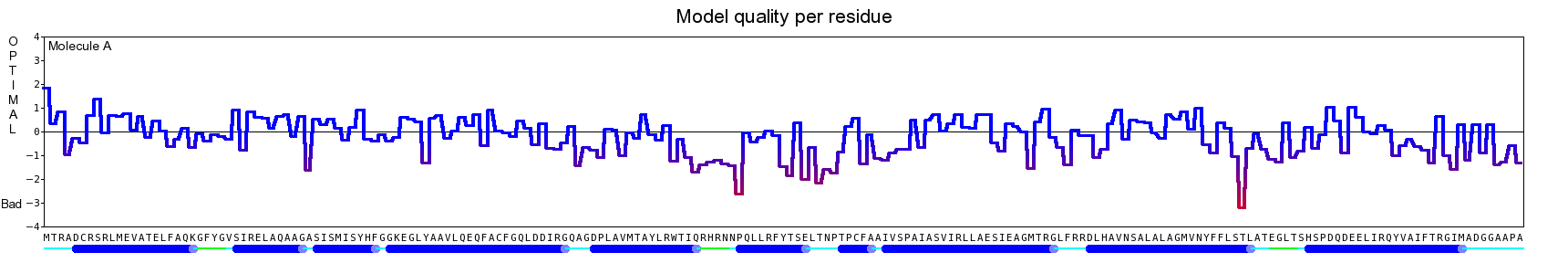 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: A :|:|:AT F :KG| G:S::E|A AGAS : | HF KE L|: V: : : ::| | : A :| :: :L R: :: : A : :: :| A: :| :RGL :A L GM: |: |: G : :S | E: V F
Template:TRSARKDREIIQAATAAFISKGYDGTSMEEIATKAGASKQTVYKHFTDKETLFGEVVLSTASQVNDIIESVTTLL..GLQQLARRLIAVLMDEELLKLRRLIIANADRMPQLGRAWYEKGFERMLASTASCFQKLTNRGL.....DPYLAASHLFGMLLWIPMNEAMFTGSNRRSKAELERHADASVEAFLAVY.........
SecStr: HHHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHH..HHHHHHHHHHHHHHCHHHHHHHHHHHHHCCCCHHHHHHHHHHCHHHHHHHHHHHHHHHHHHCC.....HHHHHHHHHHHHHCHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHH.........
The following 3 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AGMTR | GLFRRDL | HAVNS |
| 2 | RGQAG | DPL | AVMTA |
| 3 | FTRGI | MADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_3bhq-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.735 | Satisfactory |
| Packing 1D | -1.171 | Satisfactory |
| Packing 3D | -0.505 | Good |
| Overall | -0.943 | Good |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_3bhq-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.257 | Satisfactory |
| Packing 1D | -0.075 | Good |
| Packing 3D | -0.342 | Good |
| Overall | -0.370 | Good |
Since the overall quality Z-score improved to -0.370 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 9 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_3bhq-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
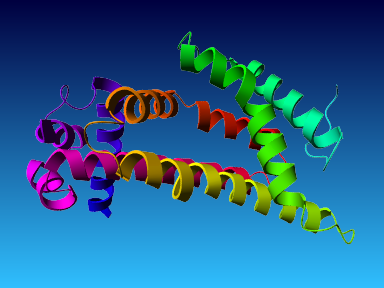 | 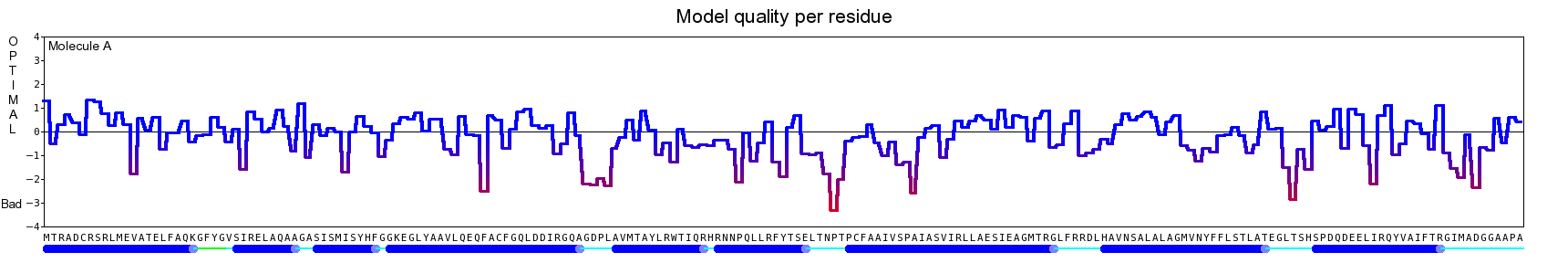 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: T A :|:|:AT F :KG| G:S::E|A AGAS : | HF KE L|: V: : : ::| | : A :| :: :L R: :: : A : :: :| A: :| :RGL :A L GM: |: |: G : :S | E: V F
Template:EtSARKDREIIQAATAAFISKGYDGTSMEEIATKAGASKQTVYKHFTDKETLFGEVVLSTASQVNDIIESVTTLL..GLQQLARRLIAVLMDEELLKLRRLIIANADRMPQLGRAWYEKGFERMLASTASCFQKLTNRGL.....DPYLAASHLFGMLLWIPMNEAMFTGSNRRSKAELERHADASVEAFLAVY.........
SecStr: CHHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHH..HHHHHHHHHHHHHHCHHHHHHHHHHHHHCCCCHHHHHHHHHHCHHHHHHHHHHHHHHHHHHCC.....HHHHHHHHHHHHHCHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHH.........
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AGMTR | GLFRRDL | HAVNS |
| 2 | MT | R | ADCRS |
| 3 | RGQAG | DPL | AVMTA |
| 4 | FTRGI | MADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_3bhq-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.899 | Satisfactory |
| Packing 1D | -1.305 | Satisfactory |
| Packing 3D | -0.692 | Good |
| Overall | -1.106 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_3bhq-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.253 | Satisfactory |
| Packing 1D | -0.289 | Good |
| Packing 3D | -0.469 | Good |
| Overall | -0.512 | Good |
Since the overall quality Z-score improved to -0.512 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 9 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_3bhq-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 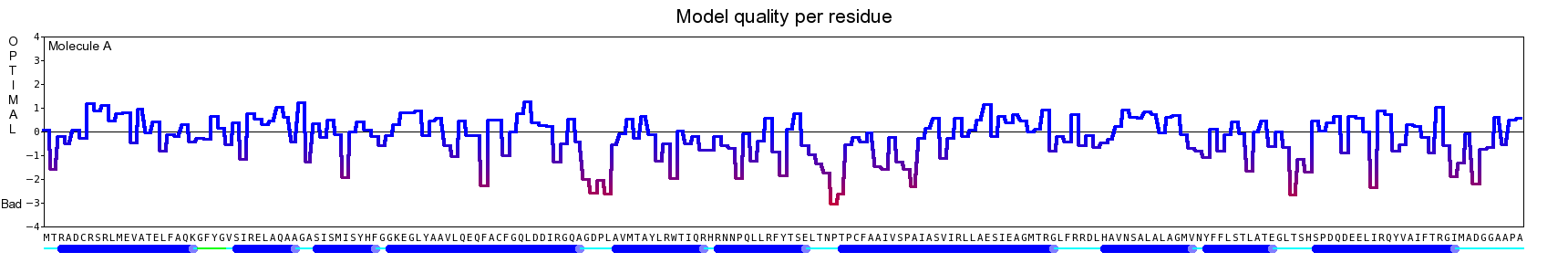 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: A :|:|:AT F :KG| G:S::E|A AGAS : | HF KE L|: V: : : ::| | ::: A :| :: :L R: :: : A : :: :| A: :| :RGL :A L GM: |: |: G : :S | E: V F
Template:TRSARKDREIIQAATAAFISKGYDGTSMEEIATKAGASKQTVYKHFTDKETLFGEVVLSTASQVNDIIESVT...MEGGlQLARRLIAVLMDEELLKLRRLIIANADRMPQLGRAWYEKGFERMLASTASCFQKLTNRGL.....DPYLAASHLFGMLLWIPMNEAMFTGSNRRSKAELERHADASVEAFLAVY.........
SecStr: HHHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHH...HHHHHHHHHHHHHHHHCHHHHHHHHHHHHHCCCCHHHHHHHHHHCHHHHHHHHHHHHHHHHHHCC.....HHHHHHHHHHHHHCHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHH.........
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AGMTR | GLFRRDL | HAVNS |
| 2 | DDIRG | QAG | DPLAV |
| 3 | GDPLA | V | MTAYL |
| 4 | FTRGI | MADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_3bhq-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.554 | Satisfactory |
| Packing 1D | -1.428 | Satisfactory |
| Packing 3D | -0.483 | Good |
| Overall | -1.007 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_3bhq-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.046 | Satisfactory |
| Packing 1D | -0.368 | Good |
| Packing 3D | -0.180 | Good |
| Overall | -0.379 | Good |
Since the overall quality Z-score improved to -0.379 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 9 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_3bhq-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 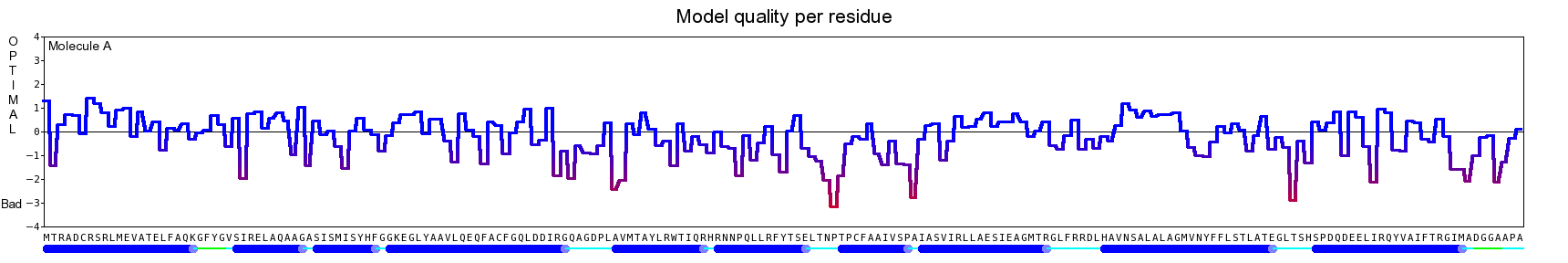 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| |:P : : :: L E || G :G:F: : : :: : : D L| : |: :T
Template:.SREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEII.....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVL.............PKFKKFID..LVSRIISALTD...........
SecStr: .HHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHCTTTTCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHH.....HHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHH.............HHHHHHHH..HHHHHHHHHHH...........
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGL | TSHSP |
| 2 | SPDQD | EE | LIRQY |
| 3 | TSELT | NPTPC | FAAIV |
| 4 | None | M | TRADC |
| 5 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fx0-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.925 | Satisfactory |
| Packing 1D | -2.155 | Poor |
| Packing 3D | -1.927 | Satisfactory |
| Overall | -2.016 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fx0-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.904 | Good |
| Packing 1D | -0.889 | Good |
| Packing 3D | -1.091 | Satisfactory |
| Overall | -0.985 | Good |
Since the overall quality Z-score improved to -0.985 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 12 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fx0-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 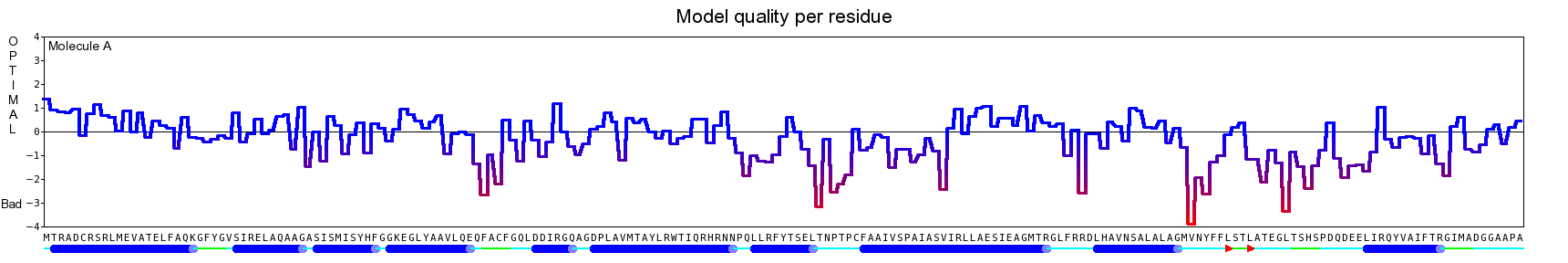 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| |:P : : :: L E || G :G:F: : : :: F : | L| : |: :T
Template:.SREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEII.....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVLFPKFKKFID..............LVSRIISALTD...........
SecStr: .HHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHCTTTTCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHH.....HHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHCHHHHHHHH..............HHHHHHHHHHH...........
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LSTLA | TEGLTSHSPDQDEE | LIRQY |
| 2 | TSELT | NPTPC | FAAIV |
| 3 | None | M | TRADC |
| 4 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fx0-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.153 | Satisfactory |
| Packing 1D | -2.114 | Poor |
| Packing 3D | -1.427 | Satisfactory |
| Overall | -1.655 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fx0-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.593 | Good |
| Packing 1D | -0.816 | Good |
| Packing 3D | -0.731 | Good |
| Overall | -0.744 | Good |
Since the overall quality Z-score improved to -0.744 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 12 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fx0-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
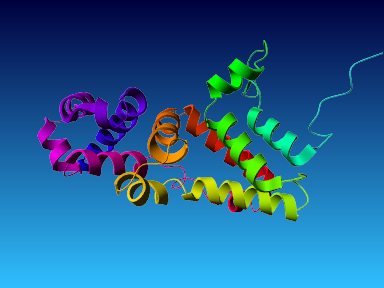 | 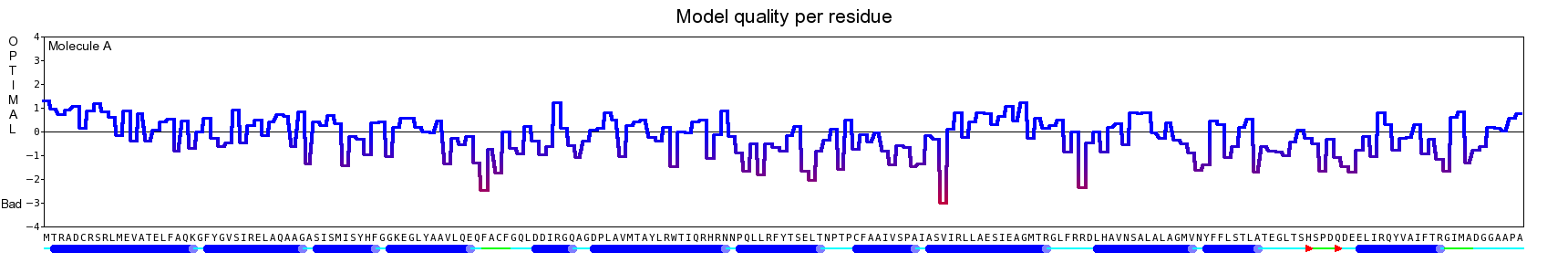 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| |:P : : :: L E || G :G:F: : : :: : | L| : |: :T
Template:.SREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEII.....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVL......PKFKKFID.........LVSRIISALTD...........
SecStr: .HHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHCTTTTCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHH.....HHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHH......HHHHHHHH.........HHHHHHHHHHH...........
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLS | TLATE |
| 2 | TEGLT | SHSPDQDEE | LIRQY |
| 3 | TSELT | NPTPC | FAAIV |
| 4 | None | M | TRADC |
| 5 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fx0-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.858 | Satisfactory |
| Packing 1D | -2.187 | Poor |
| Packing 3D | -1.909 | Satisfactory |
| Overall | -2.010 | Poor |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fx0-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.765 | Good |
| Packing 1D | -0.921 | Good |
| Packing 3D | -0.961 | Good |
| Overall | -0.917 | Good |
Since the overall quality Z-score improved to -0.917 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 12 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fx0-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
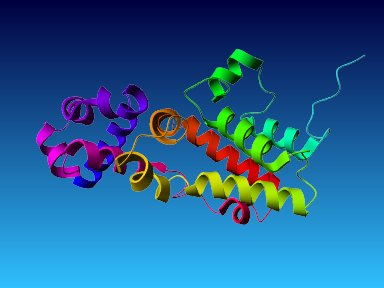 | 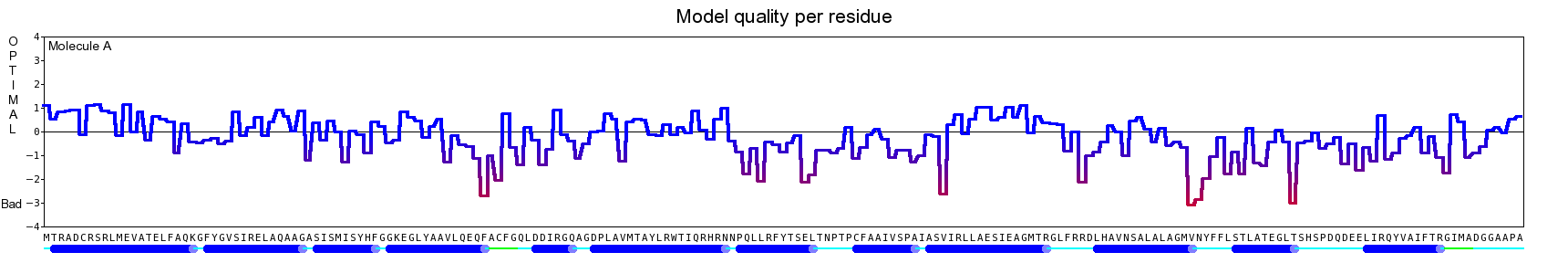 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| |:P : : :: L E || G :G:F: : : :: : D L| : |: :T
Template:.SREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEII.....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHWITSIVL...........PKFKKFID....LVSRIISALTD...........
SecStr: .HHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHCTTTTCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHH.....HHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHH...........HHHHHHHH....HHHHHHHHHHH...........
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATE | GLTSH |
| 2 | SHSPD | QDEE | LIRQY |
| 3 | TSELT | NPTPC | FAAIV |
| 4 | None | M | TRADC |
| 5 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fx0-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -2.223 | Poor |
| Packing 1D | -1.708 | Satisfactory |
| Packing 3D | -1.994 | Satisfactory |
| Overall | -1.916 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fx0-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.234 | Satisfactory |
| Packing 1D | -0.519 | Good |
| Packing 3D | -1.280 | Satisfactory |
| Overall | -0.976 | Good |
Since the overall quality Z-score improved to -0.976 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 12 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fx0-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
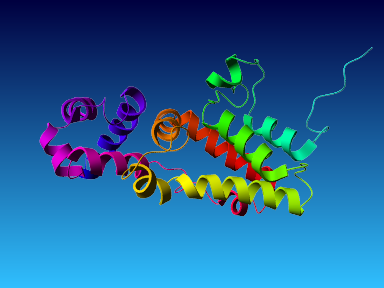 | 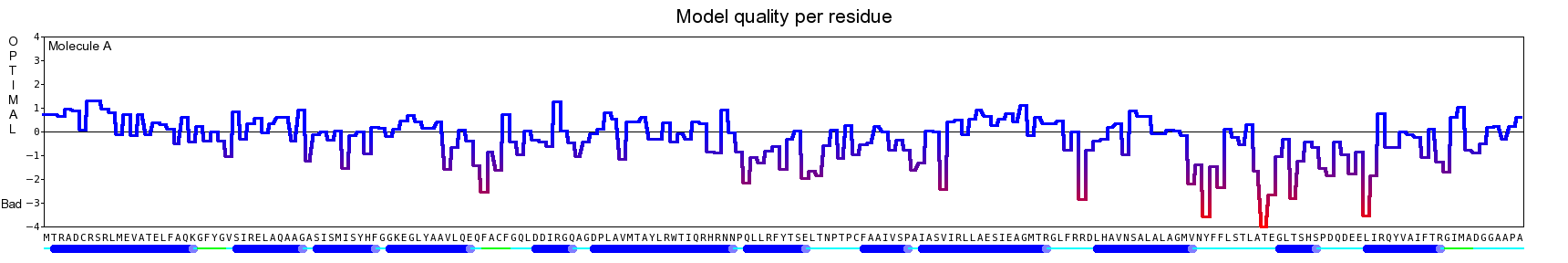 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :R : ::||::A : F:||G| G:SI:E|A: A ::|:M SY|F:GKE:LY V::: : : |: : :P| :| YL : |:NP|| : :E| |:P : : :: L E || G :G:F: : |:::V | :: : L| : |: :T
Template:.SREQTMENILKAAKKKFGERGYEGTSIQEIAKEAKVNVAMASYYFNGKENLYYEVFKKYGLANELPNFLEKNQFNPINALREYLTVFTTHIKENPEIGTLAYEEII.....RLEKIKPYFIGSFEQLKEILQEGEKQGVFHFFSINHTIHW.ITSIVLFPKFKKFID.............LVSRIISALTD...........
SecStr: .HHHHHHHHHHHHHHHHHHHHTTTTCHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHCTTTTCCHHHHHCCCHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHH.....HHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHH.HHHHHHCHHHHHHHH.............HHHHHHHHHHH...........
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | STLAT | EGLTSHSPDQDEE | LIRQY |
| 2 | VNSAL | AL | AGMVN |
| 3 | TSELT | NPTPC | FAAIV |
| 4 | None | M | TRADC |
| 5 | AIFTR | GIMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2fx0-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.689 | Satisfactory |
| Packing 1D | -1.933 | Satisfactory |
| Packing 3D | -1.587 | Satisfactory |
| Overall | -1.737 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2fx0-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.901 | Good |
| Packing 1D | -0.495 | Good |
| Packing 3D | -0.915 | Good |
| Overall | -0.749 | Good |
Since the overall quality Z-score improved to -0.749 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 12 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2fx0-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
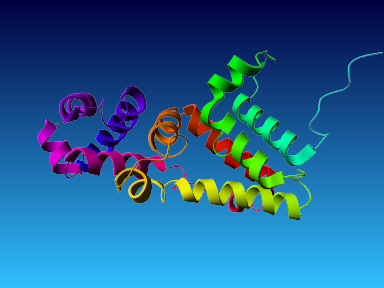 | 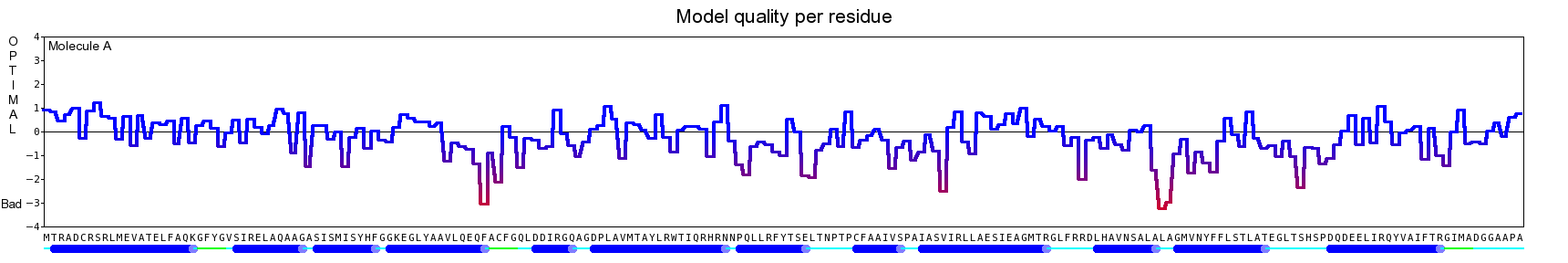 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : : |: ||::A | F:Q GF|G: |:||A| AG:S : | Y|F :KE:LY AVL:| : : | PLA:: Y|R || R: PQ R:| E| :P : :: | ::I : I : : G H | A : F :: A: G T :: :| |:: I: :GI
Template:.AVSAKKKAILSAALDTFSQFGFHGTRLEQIAELAGVSKTNLLYYFPSKEALYIAVLRQILDIWLAPLKAFREDFAPLAAIKEYIRLKLEVSRDYPQASRLFCMEMLAGAPLLMDELTGDLKALIDEKSALIAGWVKSG....PQHLIFMIWASTQHYADFAPQVEAVTGATLRDEVFFNQTVENVQRIIIEGI.........
SecStr: .HHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCCCHHHHHHCHHHHHHHHHHHHHHHHHHHC....HHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHC.........
The following 3 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AGMTR | GLFRR | DLHAV |
| 2 | None | M | TRADC |
| 3 | FTRGI | MADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_1pb6-b01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.921 | Satisfactory |
| Packing 1D | -2.029 | Poor |
| Packing 3D | -1.598 | Satisfactory |
| Overall | -1.813 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_1pb6-b01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.029 | Satisfactory |
| Packing 1D | -0.571 | Good |
| Packing 3D | -0.507 | Good |
| Overall | -0.608 | Good |
Since the overall quality Z-score improved to -0.608 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 10 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_1pb6-b01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
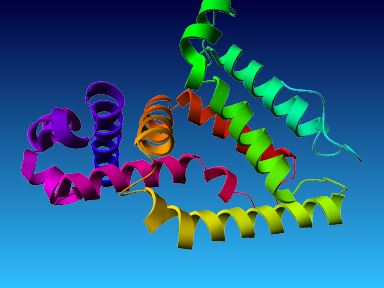 | 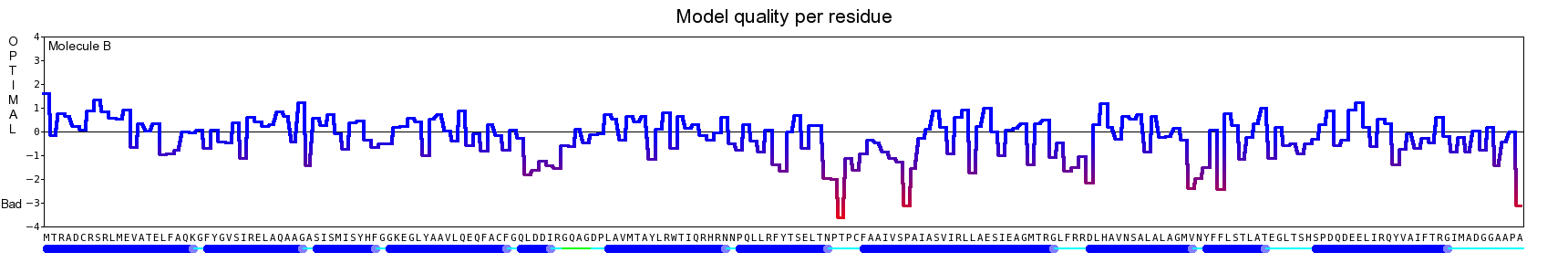 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: :::|: ||::A | F:Q GF|G: |:||A| AG:S : | Y|F :KE:LY AVL:| : : | PLA:: Y|R || R: PQ R:| E| :P : :: | ::I : I : : G H | A : F :: A: G T :: :| |:: I: :GI
Template:..AVSaKKAILSAALDTFSQFGFHGTRLEQIAELAGVSKTNLLYYFPSKEALYIAVLRQILDIWLAPLKAFREDFAPLAAIKEYIRLKLEVSRDYPQASRLFCMEMLAGAPLLMDELTGDLKALIDEKSALIAGWVKSG....PQHLIFMIWASTQHYADFAPQVEAVTGATLRDEVFFNQTVENVQRIIIEGI.........
SecStr: ..HHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHCCCCCHHHHHHCHHHHHHHHHHHHHHHHHHHC....HHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHC.........
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | AGMTR | GLFRR | DLHAV |
| 2 | MTRAD | C | RSRLM |
| 3 | None | MT | RADCR |
| 4 | FTRGI | MADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_1pb6-b02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -2.025 | Poor |
| Packing 1D | -2.020 | Poor |
| Packing 3D | -1.679 | Satisfactory |
| Overall | -1.862 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_1pb6-b02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.102 | Satisfactory |
| Packing 1D | -0.635 | Good |
| Packing 3D | -0.728 | Good |
| Overall | -0.746 | Good |
Since the overall quality Z-score improved to -0.746 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 11 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_1pb6-b02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
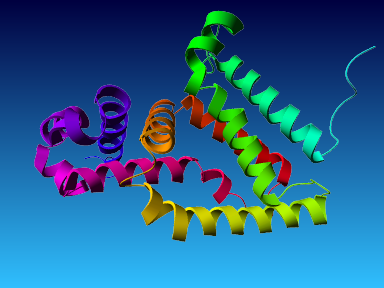 | 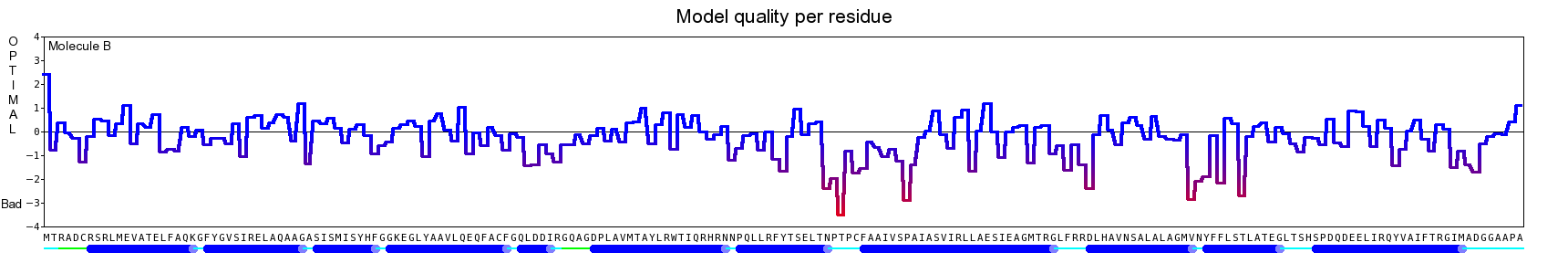 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |: R::L|||A: LFA||G: :::|R||A:AAG : | |HF :KE:| |L: : : : D A:| A : : | :: | : : F : :S :: :L |EAG: G FR D| : : L : | : |QY::I G A:P
Template:.GKSGRRTELLDIAATLFAERGLRATTVRDIADAAGILSGSLYHHFDSKESMVDEILRGFLDDLFGKYREIVAS.DSRATLEALVTTSYEAIdHSAVAIYQDEVKHLVANERF.TYLSELNTEFRELWMGVLEAGVKDGSFRSDIDVELAFRFLRDTA...................TVDTVAKQYLSIVLDG......ASP.
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHHHHCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHHHHHHHHHHHHHHHHHHHHHHCTTTTCCTTTT.HHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHH...................HHHHHHHHHHHHHHHC......CCC.
The following 7 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGLTSHSPD | QDEEL |
| 2 | YTSEL | TNPTPCFA | AIVSP |
| 3 | TIQRH | R | NNPQL |
| 4 | IRGQA | GDP | LAVMT |
| 5 | IFTRG | IMADGGA | APA |
| 6 | None | M | TRADC |
| 7 | GGAAP | A | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ibd-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.968 | Good |
| Packing 1D | -1.896 | Satisfactory |
| Packing 3D | -0.861 | Good |
| Overall | -1.281 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ibd-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.598 | Good |
| Packing 1D | -1.072 | Satisfactory |
| Packing 3D | -0.655 | Good |
| Overall | -0.809 | Good |
Since the overall quality Z-score improved to -0.809 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 2 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ibd-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
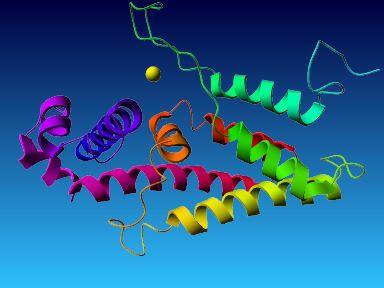 | 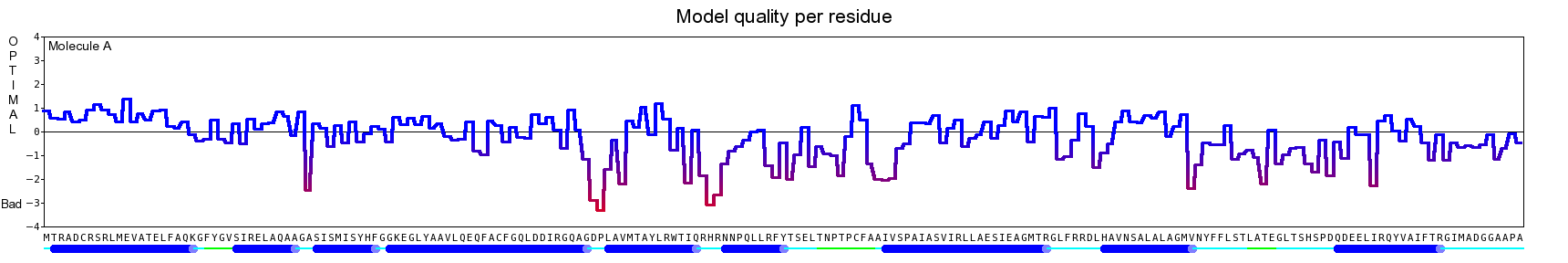 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |: R::L|||A: LFA||G: :::|R||A:AAG : | |HF :KE:| |L: : : : D A:| A : : | :: | : : :S :: :L |EAG: G FR D| : : L : | : |QY::I G
Template:.GKSGRRTELLDIAATLFAERGLRATTVRDIADAAGILSGSLYHHFDSKESMVDEILRGFLDDLFGKYREIVAS.DSRATLEALVTTSYEA.HSAVAIYQD.............TYLSELNTEFRELWMGVLEAGVKDGSFRSDIDVELAFRFLRDTA...................TVDTVAKQYLSIVLDG..........
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHHHHCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHHHHHHHHHHH.HHHHHHHHH.............HHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHH...................HHHHHHHHHHHHHHHC..........
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGLTSHSPD | QDEEL |
| 2 | QLLRF | YTSELTNPTPCFA | AIVSP |
| 3 | IRGQA | GDP | LAVMT |
| 4 | WTIQR | H | RNNPQ |
| 5 | None | M | TRADC |
| 6 | IFTRG | IMADGGAAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ibd-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.586 | Good |
| Packing 1D | -2.068 | Poor |
| Packing 3D | -0.633 | Good |
| Overall | -1.186 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ibd-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.193 | Good |
| Packing 1D | -1.311 | Satisfactory |
| Packing 3D | -0.386 | Good |
| Overall | -0.719 | Good |
Since the overall quality Z-score improved to -0.719 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 11 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ibd-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
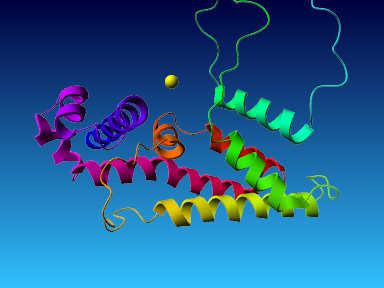 | 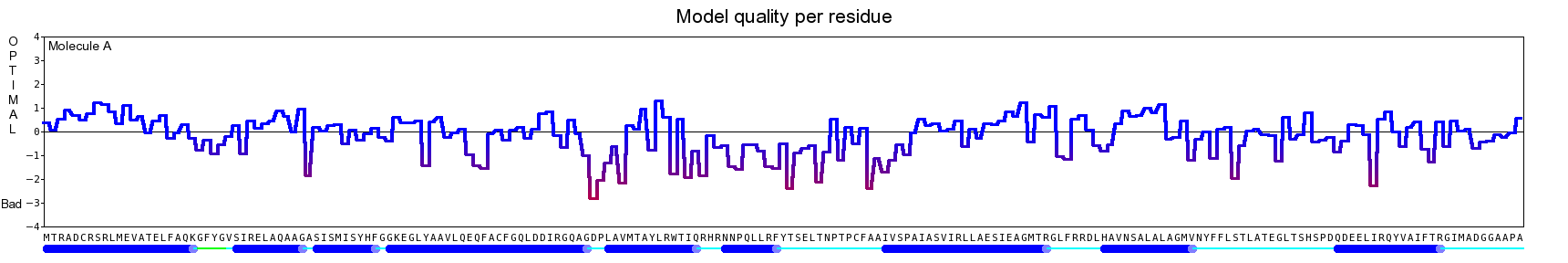 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |: R::L|||A: LFA||G: :::|R||A:AAG : | |HF :KE:| |L: : : : D A:| A : : | : : : :Y : :S :: :L |EAG: G FR D| : : L : | : |QY::I G A:P
Template:.GKSGRRTELLDIAATLFAERGLRATTVRDIADAAGILSGSLYHHFDSKESMVDEILRGFLDDLFGKYREIVAS.DSRATLEALVTTSYEAIDASHSAVAIY............TYLSELNTEFRELWMGVLEAGVKDGSFRSDIDVELAFRFLRDTA...................TVDTVAKQYLSIVLDG......ASP.
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHHHHCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHHHHHHHHHHHHHHCHHHHHHH............HHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHH...................HHHHHHHHHHHHHHHC......CCC.
The following 6 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGLTSHSPD | QDEEL |
| 2 | LLRFY | TSELTNPTPCFA | AIVSP |
| 3 | IRGQA | GDP | LAVMT |
| 4 | IFTRG | IMADGGA | APA |
| 5 | None | M | TRADC |
| 6 | GGAAP | A | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ibd-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.784 | Good |
| Packing 1D | -1.905 | Satisfactory |
| Packing 3D | -0.784 | Good |
| Overall | -1.222 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ibd-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.546 | Good |
| Packing 1D | -0.908 | Good |
| Packing 3D | -0.497 | Good |
| Overall | -0.665 | Good |
Since the overall quality Z-score improved to -0.665 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 2 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ibd-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
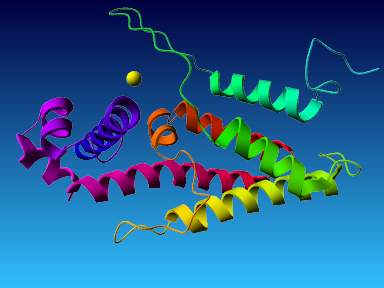 | 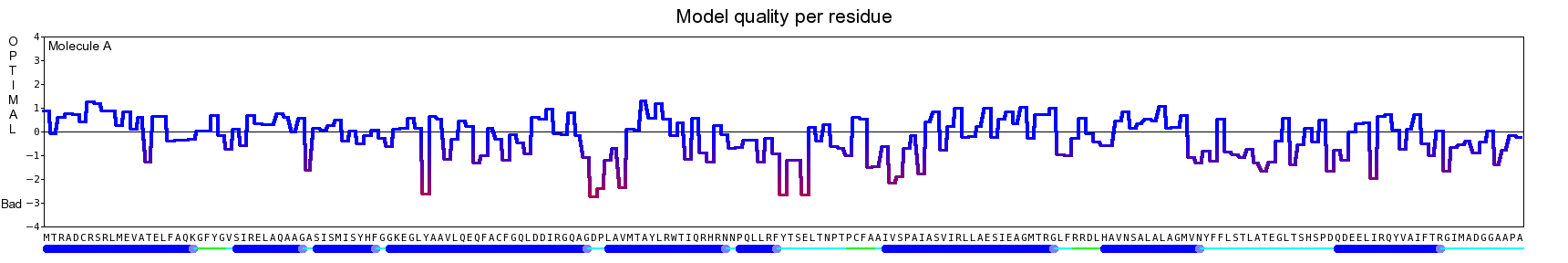 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |: R::L|||A: LFA||G: :::|R||A:AAG : | |HF :KE:| |L: : : : D A:| A : : | :: | : : : F : :S :: :L |EAG: G FR D| : : L : | : |QY::I G A:P
Template:.GKSGRRTELLDIAATLFAERGLRATTVRDIADAAGILSGSLYHHFDSKESMVDEILRGFLDDLFGKYREIVAS.DSRATLEALVTTSYEA.HSAVAIYQ.DEVKHLVANERF.TYLSELNTEFRELWMGVLEAGVKDGSFRSDIDVELAFRFLRDTA...................TVDTVAKQYLSIVLDG......ASP.
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHHHHCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHHHHHHHHHHH.HHHHHHHH.HCTTTTCCTTTT.HHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHH...................HHHHHHHHHHHHHHHC......CCC.
The following 8 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGLTSHSPD | QDEEL |
| 2 | YTSEL | TNPTPCFA | AIVSP |
| 3 | IRGQA | GDP | LAVMT |
| 4 | WTIQR | H | RNNPQ |
| 5 | PQLLR | FY | TSELT |
| 6 | IFTRG | IMADGGA | APA |
| 7 | None | M | TRADC |
| 8 | GGAAP | A | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ibd-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.816 | Good |
| Packing 1D | -2.159 | Poor |
| Packing 3D | -0.641 | Good |
| Overall | -1.259 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ibd-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.434 | Good |
| Packing 1D | -1.333 | Satisfactory |
| Packing 3D | -0.554 | Good |
| Overall | -0.840 | Good |
Since the overall quality Z-score improved to -0.840 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 2 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ibd-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
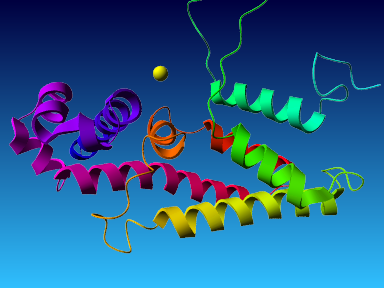 | 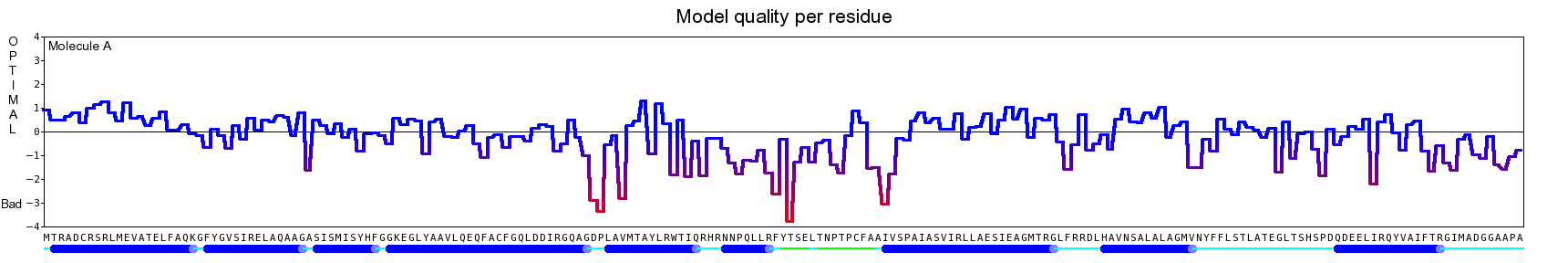 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: |: R::L|||A: LFA||G: :::|R||A:AAG : | |HF :KE:| |L: : : : D A:| A : : | :: | : : F: | : :: :L |EAG: G FR D| : : L : | : |QY::I G A:P
Template:.GKSGRRTELLDIAATLFAERGLRATTVRDIADAAGILSGSLYHHFDSKESMVDEILRGFLDDLFGKYREIVAS.DSRATLEALVTTSYEAIdHSAVAIYQDEVKHLVANERFTYLSE.LNTEFRELWMGVLEAGVKDGSFRSDIDVELAFRFLRDTA...................TVDTVAKQYLSIVLDG......ASP.
SecStr: .HHHHHHHHHHHHHHHHHHHHHTTTTHHHHHHHHHCCHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHHHHHHHHHHHHHHHHHHHHHHCTTTTCCTTTTHHHHH.HHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHH...................HHHHHHHHHHHHHHHC......CCC.
The following 7 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | LAGMV | NYFFLSTLATEGLTSHSPD | QDEEL |
| 2 | AAIVS | PA | IASVI |
| 3 | TIQRH | R | NNPQL |
| 4 | IRGQA | GDP | LAVMT |
| 5 | IFTRG | IMADGGA | APA |
| 6 | None | M | TRADC |
| 7 | GGAAP | A | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2ibd-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.061 | Satisfactory |
| Packing 1D | -1.451 | Satisfactory |
| Packing 3D | -0.868 | Good |
| Overall | -1.123 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2ibd-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.798 | Good |
| Packing 1D | -0.519 | Good |
| Packing 3D | -0.683 | Good |
| Overall | -0.636 | Good |
Since the overall quality Z-score improved to -0.636 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 2 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2ibd-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
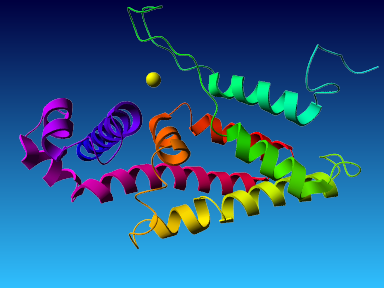 | 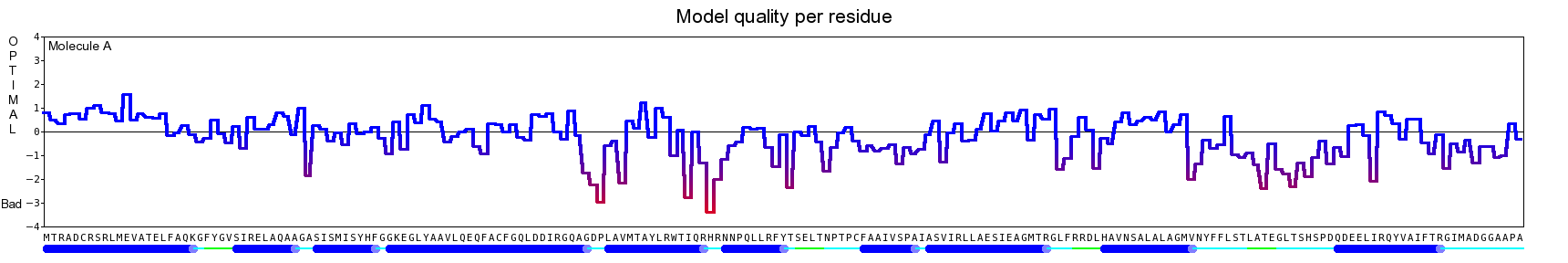 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||::A:E:F::KG| :: E|A| AG:: :|I ||F :KE LY : :: : : | : : |: RW :| : P| F : :: : | | : R:: | |: | : :D: : L : : F L T : :D : | | V |: R |
Template:....SKRDAILKAAVEVFGKKGYDRATTDEIAEKAGVAKGLIFHYFKNKEELYYQAYMSVTEKLQKEFENFLMKNRNRDIFDFMERWIEKKLEY.PEEADFLITLVSVDEG.LRKRILLDLEKSQRVFFDFVREKLKDLDLAEDVTEEIALKFLMWFFSGFEEVYLRTYQGKPELLKRDMNTLVEEVKVMLRILKKGMT....
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHH.HHHHHHHHTTTTCHHH.HHHHHHHHHHHHCHHHHHHHHHHHCCCCCTTTTHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHH....
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | QRHRN | NPQ | LLRFY |
| 2 | TNPTP | CF | AAIVS |
| 3 | None | MTRA | DCRSR |
| 4 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2iek-a01_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.121 | Satisfactory |
| Packing 1D | -2.008 | Poor |
| Packing 3D | -0.505 | Good |
| Overall | -1.181 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2iek-a01_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.484 | Good |
| Packing 1D | -0.873 | Good |
| Packing 3D | -0.134 | Good |
| Overall | -0.473 | Good |
Since the overall quality Z-score improved to -0.473 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 8 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2iek-a01.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
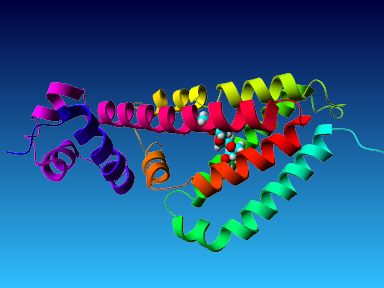 | 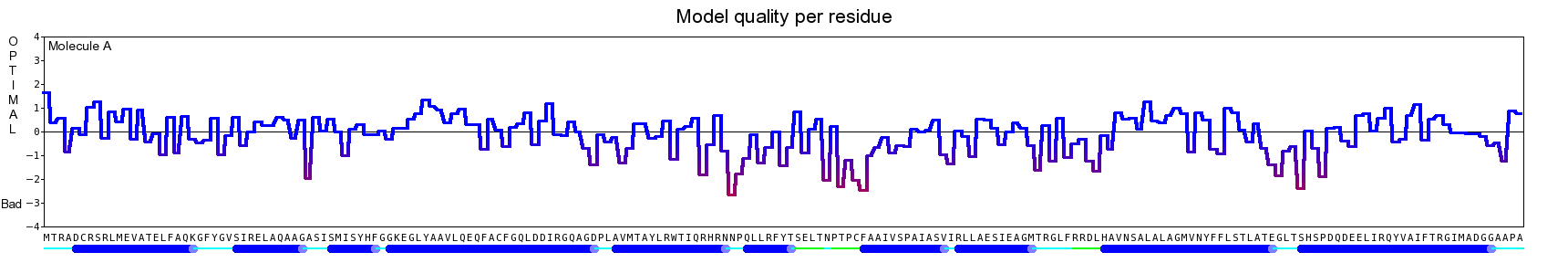 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||::A:E:F::KG| :: E|A| AG:: :|I ||F :KE LY : :: : : | : : |: RW :| : P| F : | | : R:: | |: | : :D: : L : : F L T : :D : | | V |: R |
Template:....SKRDAILKAAVEVFGKKGYDRATTDEIAEKAGVAKGLIFHYFKNKEELYYQAYMSVTEKLQKEFENFLMKNRNRDIFDFMERWIEKKLEY.PEEADFLI......DEGLRKRILLDLEKSQRVFFDFVREKLKDLDLAEDVTEEIALKFLMWFFSGFEEVYLRTYQGKPELLKRDMNTLVEEVKVMLRILKKGMT....
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHH.HHHHHHHH......HHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCTTTTHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHH....
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | QRHRN | NPQ | LLRFY |
| 2 | LRFYT | SELTNP | TPCFA |
| 3 | None | MTRA | DCRSR |
| 4 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2iek-a02_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.021 | Satisfactory |
| Packing 1D | -2.048 | Poor |
| Packing 3D | -0.457 | Good |
| Overall | -1.160 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2iek-a02_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.259 | Good |
| Packing 1D | -1.044 | Satisfactory |
| Packing 3D | -0.128 | Good |
| Overall | -0.504 | Good |
Since the overall quality Z-score improved to -0.504 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 8 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2iek-a02.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
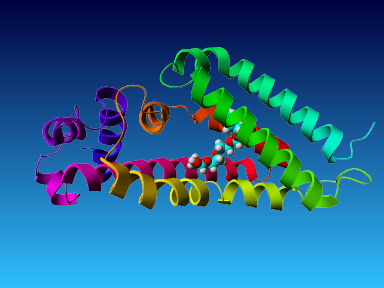 | 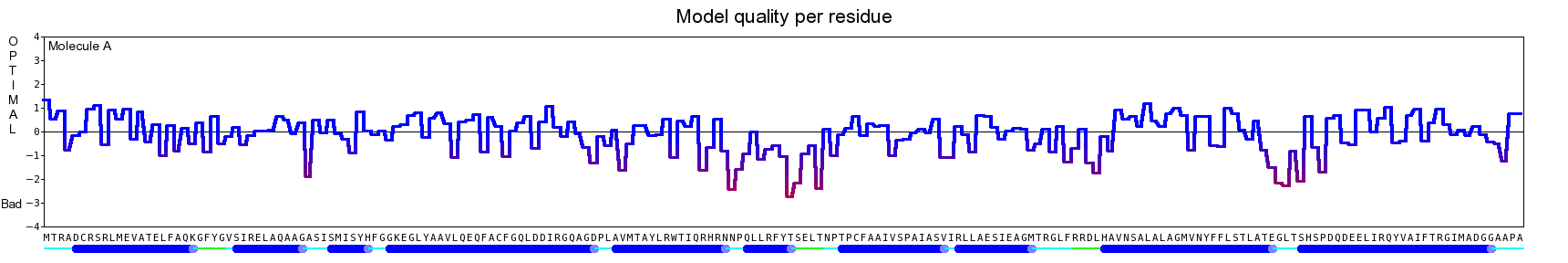 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||::A:E:F::KG| :: E|A| AG:: :|I ||F :KE LY : :: : : | : : |: RW :| : P| F : :: : | | : R:: | |: | : :D: : L : : F L T : :D : | | V |: R I| G :
Template:....SKRDAILKAAVEVFGKKGYDRATTDEIAEKAGVAKGLIFHYFKNKEELYYQAYMSVTEKLQKEFENFLMKNRNRDIFDFMERWIEKKLEY.PEEADFLITLVSVDEG.LRKRILLDLEKSQRVFFDFVREKLKDLDLAEDVTEEIALKFLMWFFSGFEEVYLRTYQGKPELLKRDMNTLVEEVKVMLR.ILKKGMT...
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHH.HHHHHHHHTTTTCHHH.HHHHHHHHHHHHCHHHHHHHHHHHCCCCCTTTTHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHH.HHHHHHH...
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | QRHRN | NPQ | LLRFY |
| 2 | TNPTP | CF | AAIVS |
| 3 | VAIFT | RG | IMADG |
| 4 | ADGGA | APA | None |
| 5 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2iek-a03_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.153 | Satisfactory |
| Packing 1D | -2.133 | Poor |
| Packing 3D | -0.553 | Good |
| Overall | -1.257 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2iek-a03_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.576 | Good |
| Packing 1D | -1.125 | Satisfactory |
| Packing 3D | -0.266 | Good |
| Overall | -0.646 | Good |
Since the overall quality Z-score improved to -0.646 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 7 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2iek-a03.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 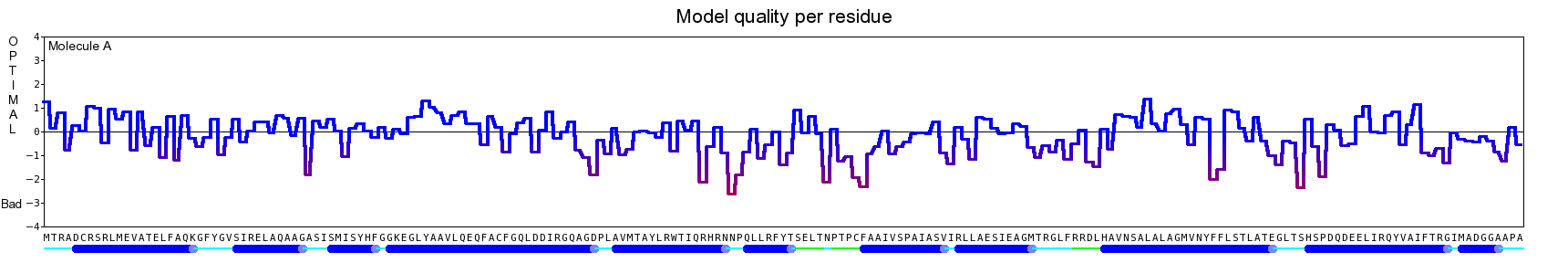 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||::A:E:F::KG| :: E|A| AG:: :|I ||F :KE LY : :: : : | : : |: RW :| : P| F : | | : R:: | |: | : :D: : L : : F L T : :D : | | V |: R I| G :
Template:....SKRDAILKAAVEVFGKKGYDRATTDEIAEKAGVAKGLIFHYFKNKEELYYQAYMSVTEKLQKEFENFLMKNRNRDIFDFMERWIEKKLEY.PEEADFLI......DEGLRKRILLDLEKSQRVFFDFVREKLKDLDLAEDVTEEIALKFLMWFFSGFEEVYLRTYQGKPELLKRDMNTLVEEVKVMLR.ILKKGMT...
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHH.HHHHHHHH......HHHHHHHHHHHHHHHCHHHHHHHHHHHCCCCCTTTTHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHH.HHHHHHH...
The following 5 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | QRHRN | NPQ | LLRFY |
| 2 | VAIFT | RG | IMADG |
| 3 | LRFYT | SELTNP | TPCFA |
| 4 | ADGGA | APA | None |
| 5 | None | MTRA | DCRSR |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2iek-a04_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.044 | Satisfactory |
| Packing 1D | -2.279 | Poor |
| Packing 3D | -0.628 | Good |
| Overall | -1.333 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2iek-a04_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.428 | Good |
| Packing 1D | -1.163 | Satisfactory |
| Packing 3D | -0.305 | Good |
| Overall | -0.658 | Good |
Since the overall quality Z-score improved to -0.658 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 7 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2iek-a04.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
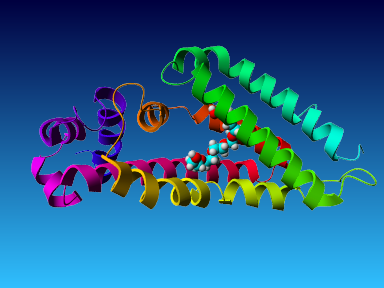 | 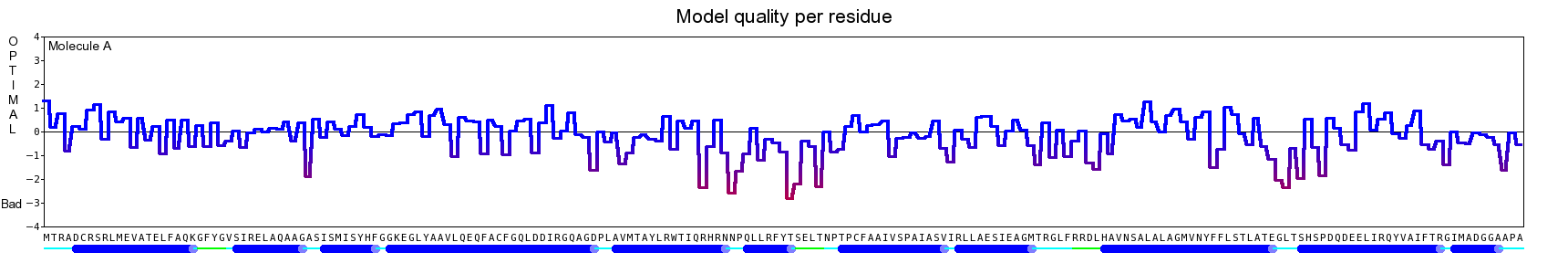 |
This model is a monomer, and based on the following alignment:
SecStr: CCCCCHHHHHHHHHHHHHHHHHHCCEEEHHHHHHHCCCEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHCCCCHHHHHEECCCCCCCCCCHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHEHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHCCCCCCCCCCC
Target: MTRADCRSRLMEVATELFAQKGFYGVSIRELAQAAGASISMISYHFGGKEGLYAAVLQEQFACFGQLDDIRGQAGDPLAVMTAYLRWTIQRHRNNPQLLRFYTSELTNPTPCFAAIVSPAIASVIRLLAESIEAGMTRGLFRRDLHAVNSALALAGMVNYFFLSTLATEGLTSHSPDQDEELIRQYVAIFTRGIMADGGAAPA
Match: : R: ||::A:E:F::KG| :: E|A| AG:: :|I ||F :KE LY : :: : : | : : |: RW :| : P| F : :: I: S R:: | |: | : :D: : L : : F L T : :D : | | V |: R |
Template:....SKRDAILKAAVEVFGKKGYDRATTDEIAEKAGVAKGLIFHYFKNKEELYYQAYMSVTEKLQKEFENFLMKNRNRDIFDFMERWIEKKLEY.PEEADFLITLVSVDEGLRKRILLDLEKS..RVFFDFVREKLKDLDLAEDVTEEIALKFLMWFFSGFEEVYLRTYQGKPELLKRDMNTLVEEVKVMLRILKKGMT....
SecStr: ....HHHHHHHHHHHHHHHHHCTTTTHHHHHHHHHHCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHH.HHHHHHHHTTTTCHHHHHHHHHHHHHHH..HHHHHHHHHHHCCCCCTTTTHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHH....
The following 4 loops had to be modeled:
| Loop | N-terminal anchor | Loop sequence | C-terminal anchor |
| 1 | PAIAS | VI | RLLAE |
| 2 | QRHRN | NPQ | LLRFY |
| 3 | None | MTRA | DCRSR |
| 4 | MADGG | AAPA | None |
After the side-chains had been built, optimized and fine-tuned, all newly modeled parts were subjected to a combined steepest descent and simulated annealing minimization (i.e. the backbone atoms of aligned residues were kept fixed to avoid potential damage).
The resulting half-refined model has been saved as t0454_2iek-a05_refined050.yob and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -1.066 | Satisfactory |
| Packing 1D | -2.071 | Poor |
| Packing 3D | -0.404 | Good |
| Overall | -1.151 | Satisfactory |
Then a full unrestrained simulated annealing minimization was run for the entire model. The result has been saved as t0454_2iek-a05_refined100.yob, the corresponding Z-scores are listed below:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.508 | Good |
| Packing 1D | -0.924 | Good |
| Packing 3D | -0.113 | Good |
| Overall | -0.487 | Good |
Since the overall quality Z-score improved to -0.487 during the minimization, this fully refined model has been accepted as the final one for this template and alignment.
NOTE: 8 residues contained in N- and C-terminal floppy tails of the monomeric model have been excluded from the Z-scores in both tables above, since they would have mainly added noise to the result.
The final model for this template and alignment has been saved as t0454_2iek-a05.yob, and is shown below, together with a plot of its average per-residue quality Z-scores:
 | 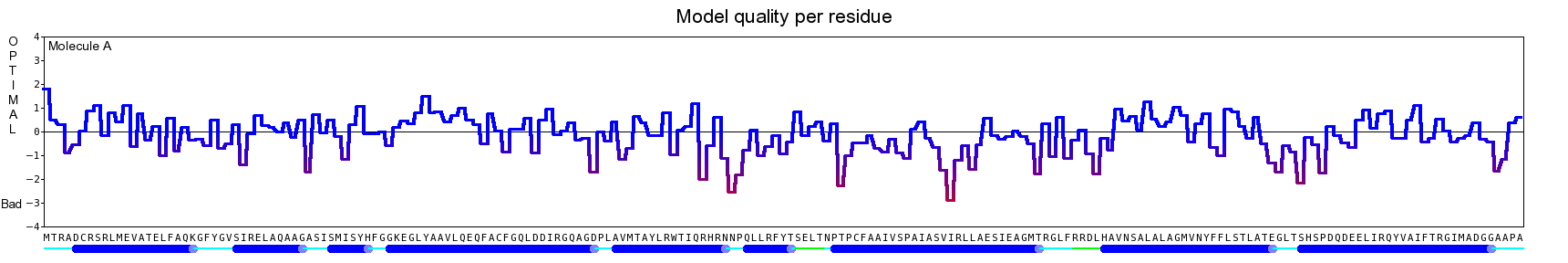 |
The following table lists the 42 models sorted by their overall quality Z-scores. The models have been superposed and saved together as t0454.sce.
| Rank | Z-score | Structure | State | Model ID | Filename | Original number | Comment |
| 1 | -0.332 | monomeric | BFBQ-A03 | t0454_bfbq-a03.yob | 15 | Good | |
| 2 | -0.370 | monomeric | 3BHQ-A01 | t0454_3bhq-a01.yob | 23 | Good | |
| 3 | -0.379 | monomeric | 3BHQ-A03 | t0454_3bhq-a03.yob | 25 | Good | |
| 4 | -0.473 | monomeric | 2IEK-A01 | t0454_2iek-a01.yob | 38 | Good | |
| 5 | -0.487 | monomeric | 2IEK-A05 | t0454_2iek-a05.yob | 42 | Good | |
| 6 | -0.504 | monomeric | 2IEK-A02 | t0454_2iek-a02.yob | 39 | Good | |
| 7 | -0.512 | monomeric | 3BHQ-A02 | t0454_3bhq-a02.yob | 24 | Good | |
| 8 | -0.532 | monomeric | BFBQ-A02 | t0454_bfbq-a02.yob | 14 | Good | |
| 9 | -0.548 | monomeric | 2EH3-A01 | t0454_2eh3-a01.yob | 18 | Good | |
| 10 | -0.578 | monomeric | 2FBQ-A03 | t0454_2fbq-a03.yob | 10 | Good | |
| 11 | -0.600 | monomeric | 2EH3-A05 | t0454_2eh3-a05.yob | 22 | Good | |
| 12 | -0.608 | monomeric | 1PB6-B01 | t0454_1pb6-b01.yob | 31 | Good | |
| 13 | -0.627 | monomeric | BFBQ-A04 | t0454_bfbq-a04.yob | 16 | Good | |
| 14 | -0.636 | monomeric | 2IBD-A05 | t0454_2ibd-a05.yob | 37 | Good | |
| 15 | -0.646 | monomeric | 2IEK-A03 | t0454_2iek-a03.yob | 40 | Good | |
| 16 | -0.658 | monomeric | 2IEK-A04 | t0454_2iek-a04.yob | 41 | Good | |
| 17 | -0.660 | monomeric | BFBQ-A01 | t0454_bfbq-a01.yob | 13 | Good | |
| 18 | -0.665 | monomeric | 2IBD-A03 | t0454_2ibd-a03.yob | 35 | Good | |
| 19 | -0.679 | monomeric | 2FBQ-A02 | t0454_2fbq-a02.yob | 9 | Good | |
| 20 | -0.685 | monomeric | 2JJ7-B02 | t0454_2jj7-b02.yob | 7 | Good | |
| 21 | -0.712 | monomeric | 2JJ7-B01 | t0454_2jj7-b01.yob | 6 | Good | |
| 22 | -0.715 | monomeric | 2FBQ-A04 | t0454_2fbq-a04.yob | 11 | Good | |
| 23 | -0.719 | monomeric | 2IBD-A02 | t0454_2ibd-a02.yob | 34 | Good | |
| 24 | -0.720 | monomeric | BFBQ-A05 | t0454_bfbq-a05.yob | 17 | Good | |
| 25 | -0.744 | monomeric | 2FX0-A02 | t0454_2fx0-a02.yob | 27 | Good | |
| 26 | -0.746 | monomeric | 1PB6-B02 | t0454_1pb6-b02.yob | 32 | Good | |
| 27 | -0.749 | monomeric | 2FX0-A05 | t0454_2fx0-a05.yob | 30 | Good | |
| 28 | -0.762 | monomeric | 2FBQ-A01 | t0454_2fbq-a01.yob | 8 | Good | |
| 29 | -0.765 | monomeric | 2EH3-A04 | t0454_2eh3-a04.yob | 21 | Good | |
| 30 | -0.778 | monomeric | 2EH3-A03 | t0454_2eh3-a03.yob | 20 | Good | |
| 31 | -0.803 | monomeric | 2FBQ-A05 | t0454_2fbq-a05.yob | 12 | Good | |
| 32 | -0.809 | monomeric | 2IBD-A01 | t0454_2ibd-a01.yob | 33 | Good | |
| 33 | -0.840 | monomeric | 2IBD-A04 | t0454_2ibd-a04.yob | 36 | Good | |
| 34 | -0.855 | monomeric | 2EH3-A02 | t0454_2eh3-a02.yob | 19 | Good | |
| 35 | -0.913 | monomeric | 2OFL-B04 | t0454_2ofl-b04.yob | 4 | Good | |
| 36 | -0.917 | monomeric | 2FX0-A03 | t0454_2fx0-a03.yob | 28 | Good | |
| 37 | -0.976 | monomeric | 2FX0-A04 | t0454_2fx0-a04.yob | 29 | Good | |
| 38 | -0.985 | monomeric | 2FX0-A01 | t0454_2fx0-a01.yob | 26 | Good | |
| 39 | -1.007 | monomeric | 2OFL-B01 | t0454_2ofl-b01.yob | 1 | Satisfactory | |
| 40 | -1.107 | monomeric | 2OFL-B05 | t0454_2ofl-b05.yob | 5 | Satisfactory | |
| 41 | -1.194 | monomeric | 2OFL-B03 | t0454_2ofl-b03.yob | 3 | Satisfactory | |
| 42 | -1.376 | monomeric | 2OFL-B02 | t0454_2ofl-b02.yob | 2 | Satisfactory |
Finally, YASARA tried to combine the best parts of the 42 models to obtain a hybrid model, hoping to increase the accuracy beyond each of the contributors. Starting with the best scoring model t0454_bfbq-a03.yob, the following fragments were successfully copied from other models:
| Transfer | First residue | Last residue | Length | From model | Energy after transfer [kcal/mol] |
| 1 | ILE A 116 | SER A 118 | 3 | 2IEK-A04 | -21362 |
| 2 | PRO A 176 | GLN A 178 | 3 | 2EH3-A05 | -21480 |
The resulting hybrid model was subjected to a final round of simulated annealing minimization in explicit solvent and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
| Dihedrals | -0.656 | Good |
| Packing 1D | -0.223 | Good |
| Packing 3D | -0.206 | Good |
| Overall | -0.278 | Good |
The following figure shows the original best model in blue, with all parts copied from other models colored yellow.
 | 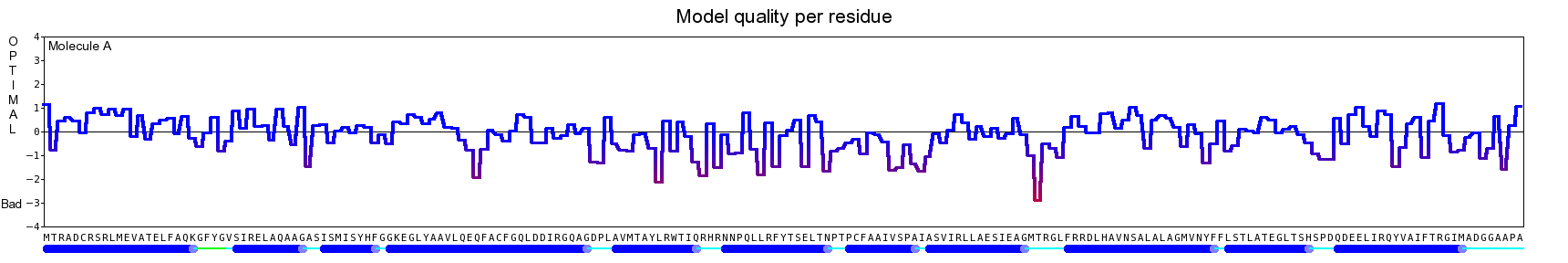 |
Since this hybrid model scored better than all previous models, it was saved as the final model t0454.yob.